A single source k-shortest paths algorithm to infer regulatory pathways in a gene network
- PMID: 22689778
- PMCID: PMC3371844
- DOI: 10.1093/bioinformatics/bts212
A single source k-shortest paths algorithm to infer regulatory pathways in a gene network
Abstract
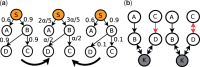
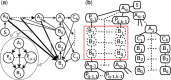
Motivation: Inferring the underlying regulatory pathways within a gene interaction network is a fundamental problem in Systems Biology to help understand the complex interactions and the regulation and flow of information within a system-of-interest. Given a weighted gene network and a gene in this network, the goal of an inference algorithm is to identify the potential regulatory pathways passing through this gene.
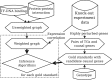
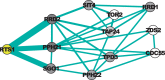
Results: In a departure from previous approaches that largely rely on the random walk model, we propose a novel single-source k-shortest paths based algorithm to address this inference problem. An important element of our approach is to explicitly account for and enhance the diversity of paths discovered by our algorithm. The intuition here is that diversity in paths can help enrich different functions and thereby better position one to understand the underlying system-of-interest. Results on the yeast gene network demonstrate the utility of the proposed approach over extant state-of-the-art inference algorithms. Beyond utility, our algorithm achieves a significant speedup over these baselines.
Availability: All data and codes are freely available upon request.
Figures




Similar articles
-
Probabilistic inference and ranking of gene regulatory pathways as a shortest-path problem.BMC Bioinformatics. 2013;14 Suppl 13(Suppl 13):S5. doi: 10.1186/1471-2105-14-S13-S5. Epub 2013 Oct 1. BMC Bioinformatics. 2013. PMID: 24266986 Free PMC article.
-
bLARS: An Algorithm to Infer Gene Regulatory Networks.IEEE/ACM Trans Comput Biol Bioinform. 2016 Mar-Apr;13(2):301-14. doi: 10.1109/TCBB.2015.2450740. IEEE/ACM Trans Comput Biol Bioinform. 2016. PMID: 27045829
-
IRIS: a method for reverse engineering of regulatory relations in gene networks.BMC Bioinformatics. 2009 Dec 23;10:444. doi: 10.1186/1471-2105-10-444. BMC Bioinformatics. 2009. PMID: 20030818 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
A review of integration strategies to support gene regulatory network construction.ScientificWorldJournal. 2012;2012:435257. doi: 10.1100/2012/435257. Epub 2012 Dec 27. ScientificWorldJournal. 2012. PMID: 23365519 Free PMC article. Review.
Cited by
-
Chapter 5: Network biology approach to complex diseases.PLoS Comput Biol. 2012;8(12):e1002820. doi: 10.1371/journal.pcbi.1002820. Epub 2012 Dec 27. PLoS Comput Biol. 2012. PMID: 23300411 Free PMC article.
-
A physarum-inspired prize-collecting steiner tree approach to identify subnetworks for drug repositioning.BMC Syst Biol. 2016 Dec 5;10(Suppl 5):128. doi: 10.1186/s12918-016-0371-3. BMC Syst Biol. 2016. PMID: 28105946 Free PMC article.
-
Detection of deregulated modules using deregulatory linked path.PLoS One. 2013 Jul 24;8(7):e70412. doi: 10.1371/journal.pone.0070412. Print 2013. PLoS One. 2013. PMID: 23894653 Free PMC article.
-
Inference on chains of disease progression based on disease networks.PLoS One. 2019 Jun 28;14(6):e0218871. doi: 10.1371/journal.pone.0218871. eCollection 2019. PLoS One. 2019. PMID: 31251766 Free PMC article.
-
Network Modeling Unravels Mechanisms of Crosstalk between Ethylene and Salicylate Signaling in Potato.Plant Physiol. 2018 Sep;178(1):488-499. doi: 10.1104/pp.18.00450. Epub 2018 Jun 22. Plant Physiol. 2018. PMID: 29934298 Free PMC article.
References
-
- Bader J.S., et al. Gaining confidence in high-throughput protein interaction networks. Nat Biotechnol. 2004;22:78–85. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

