A conditional neural fields model for protein threading
- PMID: 22689779
- PMCID: PMC3371845
- DOI: 10.1093/bioinformatics/bts213
A conditional neural fields model for protein threading
Abstract
Motivation: Alignment errors are still the main bottleneck for current template-based protein modeling (TM) methods, including protein threading and homology modeling, especially when the sequence identity between two proteins under consideration is low (<30%).
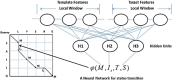
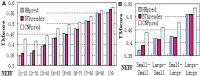
Results: We present a novel protein threading method, CNFpred, which achieves much more accurate sequence-template alignment by employing a probabilistic graphical model called a Conditional Neural Field (CNF), which aligns one protein sequence to its remote template using a non-linear scoring function. This scoring function accounts for correlation among a variety of protein sequence and structure features, makes use of information in the neighborhood of two residues to be aligned, and is thus much more sensitive than the widely used linear or profile-based scoring function. To train this CNF threading model, we employ a novel quality-sensitive method, instead of the standard maximum-likelihood method, to maximize directly the expected quality of the training set. Experimental results show that CNFpred generates significantly better alignments than the best profile-based and threading methods on several public (but small) benchmarks as well as our own large dataset. CNFpred outperforms others regardless of the lengths or classes of proteins, and works particularly well for proteins with sparse sequence profiles due to the effective utilization of structure information. Our methodology can also be adapted to protein sequence alignment.
Figures



References
-
- Akutsu T., et al. Hardness results on local multiple alignment of biological sequences. Inform. Media Technol. 2007;2:514–522.
-
- Biegert A., Söding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

