Strategy for directing combinatorial genome engineering in Escherichia coli
- PMID: 22689973
- PMCID: PMC3387050
- DOI: 10.1073/pnas.1206299109
Strategy for directing combinatorial genome engineering in Escherichia coli
Abstract
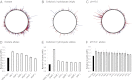
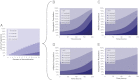
We describe a directed genome-engineering approach that combines genome-wide methods for mapping genes to traits [Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT (2010) Nat Biotechnol 28:856-862] with strategies for rapidly creating combinatorial ribosomal binding site (RBS) mutation libraries containing billions of targeted modifications [Wang HH, et al. (2009) Nature 460:894-898]. This approach should prove broadly applicable to various efforts focused on improving production of fuels, chemicals, and pharmaceuticals, among other products. We used barcoded promoter mutation libraries to map the effect of increased or decreased expression of nearly every gene in Escherichia coli onto growth in several model environments (cellulosic hydrolysate, low pH, and high acetate). Based on these data, we created and evaluated RBS mutant libraries (containing greater than 100,000,000 targeted mutations), targeting the genes identified to most affect growth. On laboratory timescales, we successfully identified a broad range of mutations (>25 growth-enhancing mutations confirmed), which improved growth rate 10-200% for several different conditions. Although successful, our efforts to identify superior combinations of growth-enhancing genes emphasized the importance of epistatic interactions among the targeted genes (synergistic, antagonistic) for taking full advantage of this approach to directed genome engineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

