Repeat associated small RNAs vary among parents and following hybridization in maize
- PMID: 22689990
- PMCID: PMC3387101
- DOI: 10.1073/pnas.1202073109
Repeat associated small RNAs vary among parents and following hybridization in maize
Abstract
Small RNAs (sRNAs) are hypothesized to contribute to hybrid vigor because they maintain genome integrity, contribute to genetic diversity, and control gene expression. We used Illumina sequencing to assess how sRNA populations vary between two maize inbred lines (B73 and Mo17) and their hybrid. We sampled sRNAs from the seedling shoot apex and the developing ear, two rapidly growing tissues that program the greater growth of maize hybrids. We found that parental differences in siRNAs primarily originate from repeat regions. Although the maize genome contains greater number and complexity of repeats compared with Arabidopsis or rice, we confirmed that, like these simpler plant genomes, 24-nt siRNAs whose abundance differs between maize parents also show a trend of down-regulation following hybridization. Surprisingly, hybrid vigor is fully maintained when 24-nt siRNAs are globally reduced by mutation of the RNA-dependent RNA polymerase 2 encoded by modifier of paramutation1 (mop1). We also discovered that 21-22-nt siRNAs derived from a number of distinct retrotransposon families differentially accumulate between B73 and Mo17 as well as their hybrid. Thus, maize possesses a unique source of genetic variation for regulating transposons and genes at a genomic scale, which may contribute to its high degree of observed heterosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

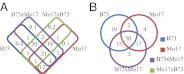
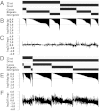


References
-
- Shull GH. The composition of field maize. American Breeding Association Reports. 1908;4:296–301.
-
- Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. - PubMed
-
- Messing J, Dooner HK. Organization and variability of the maize genome. Curr Opin Plant Biol. 2006;9:157–163. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

