The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization
- PMID: 22691236
- PMCID: PMC3488578
- DOI: 10.1186/1471-2407-12-235
The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization
Abstract
Background: Cancer-related genes show racial differences. Therefore, identification and characterization of DNA copy number alteration regions in different racial groups helps to dissect the mechanism of tumorigenesis.
Methods: Array-comparative genomic hybridization (array-CGH) was analyzed for DNA copy number profile in 40 Asian and 20 Caucasian lung cancer patients. Three methods including MetaCore analysis for disease and pathway correlations, concordance analysis between array-CGH database and the expression array database, and literature search for copy number variation genes were performed to select novel lung cancer candidate genes. Four candidate oncogenes were validated for DNA copy number and mRNA and protein expression by quantitative polymerase chain reaction (qPCR), chromogenic in situ hybridization (CISH), reverse transcriptase-qPCR (RT-qPCR), and immunohistochemistry (IHC) in more patients.
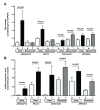
Results: We identified 20 chromosomal imbalance regions harboring 459 genes for Caucasian and 17 regions containing 476 genes for Asian lung cancer patients. Seven common chromosomal imbalance regions harboring 117 genes, included gain on 3p13-14, 6p22.1, 9q21.13, 13q14.1, and 17p13.3; and loss on 3p22.2-22.3 and 13q13.3 were found both in Asian and Caucasian patients. Gene validation for four genes including ARHGAP19 (10q24.1) functioning in Rho activity control, FRAT2 (10q24.1) involved in Wnt signaling, PAFAH1B1 (17p13.3) functioning in motility control, and ZNF322A (6p22.1) involved in MAPK signaling was performed using qPCR and RT-qPCR. Mean gene dosage and mRNA expression level of the four candidate genes in tumor tissues were significantly higher than the corresponding normal tissues (P<0.001~P=0.06). In addition, CISH analysis of patients indicated that copy number amplification indeed occurred for ARHGAP19 and ZNF322A genes in lung cancer patients. IHC analysis of paraffin blocks from Asian Caucasian patients demonstrated that the frequency of PAFAH1B1 protein overexpression was 68% in Asian and 70% in Caucasian.
Conclusions: Our study provides an invaluable database revealing common and differential imbalance regions at specific chromosomes among Asian and Caucasian lung cancer patients. Four validation methods confirmed our database, which would help in further studies on the mechanism of lung tumorigenesis.
Figures



References
-
- Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. - DOI - PubMed
-
- Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37(Suppl 8):S4–S66. - PubMed
-
- Lee LN, Shew JY, Sheu JC, Lee YC, Lee WC, Fang MT, Chang HF, Yu CJ, Yang PC, Luh KT. Exon 8 mutation of p53 gene associated with nodal metastasis in non-small-cell lung cancer. Am J Respir Crit Care Med. 1994;150:1667–1671. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

