Epigenetics of the depressed brain: role of histone acetylation and methylation
- PMID: 22692567
- PMCID: PMC3521990
- DOI: 10.1038/npp.2012.73
Epigenetics of the depressed brain: role of histone acetylation and methylation
Abstract
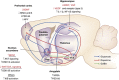
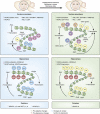
Major depressive disorder is a chronic, remitting syndrome involving widely distributed circuits in the brain. Stable alterations in gene expression that contribute to structural and functional changes in multiple brain regions are implicated in the heterogeneity and pathogenesis of the illness. Epigenetic events that alter chromatin structure to regulate programs of gene expression have been associated with depression-related behavior, antidepressant action, and resistance to depression or 'resilience' in animal models, with increasing evidence for similar mechanisms occurring in postmortem brains of depressed humans. In this review, we discuss recent advances in our understanding of epigenetic contributions to depression, in particular the role of histone acetylation and methylation, which are revealing novel mechanistic insight into the syndrome that may aid in the development of novel targets for depression treatment.
Figures

References
-
- Abe N, Uchida S, Otsuki K, Hobara T, Yamagata H, Higuchi F, et al. Altered sirtuin deacetylase gene expression in patients with a mood disorder. J Psychiatr Res. 2011;45:1106–1112. - PubMed
-
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187:591–601. - PubMed
-
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- American Psychiatric Association . Diagnostic Criteria from DSM-IV-TR. American Psychiatric Association: Washington, D.C; 2000.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

