Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan
- PMID: 22693205
- PMCID: PMC3423099
- DOI: 10.1152/ajpendo.00110.2012
Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan
Abstract
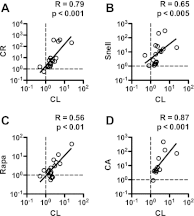
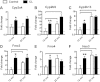
Xenobiotic metabolism has been proposed to play a role in modulating the rate of aging. Xenobiotic metabolizing enzymes (XME) are expressed at higher levels in calorically restricted mice (CR) and in GH/IGF-I-deficient, long-lived mutant mice. In this study, we show that many phase I XME genes are similarly upregulated in additional long-lived mouse models, including "crowded litter" (CL) mice, whose lifespan has been increased by food restriction limited to the first 3 wk of life, and in mice treated with rapamycin. Induction in the CL mice lasts at least through 22 mo of age, but induction by rapamycin is transient for many of the mRNAs. Cytochrome P-450s, flavin monooxygenases, hydroxyacid oxidase, and metallothioneins were found to be significantly elevated in similar proportions in each of the models of delayed aging tested, whether these were based on mutation, diet, drug treatment, or transient early intervention. The same pattern of mRNA elevation could be induced by 2 wk of treatment with tert-butylhydroquinone, an oxidative toxin known to activate Nrf2-dependent target genes. These results suggest that elevation of phase I XMEs is a hallmark of long-lived mice and may facilitate screens for agents worth testing in intervention-based lifespan studies.
Figures





References
-
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Rosenfeld SV, Blagosklonny MV. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle 10: 4230–4236, 2011 - PubMed
-
- Anwar-Mohamed A, Degenhardt OS, Gendy el MAM, Seubert JM, Kleeberger SR, El-Kadi AOS. The effect of Nrf2 knockout on the constitutive expression of drug metabolizing enzymes and transporters in C57Bl/6 mice livers. Toxicol In Vitro 25: 785–795, 2011 - PubMed
-
- Baird L, Dinkova Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol 85: 241–272, 2011 - PubMed
-
- Bartke A, Brown-Borg HM, Mattison J, Kinney B, Hauck S, Wright C. Prolonged longevity of hypopituitary dwarf mice. Exp Gerontol 36: 21–28, 2001 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

