Simulation of molecular data under diverse evolutionary scenarios
- PMID: 22693434
- PMCID: PMC3364941
- DOI: 10.1371/journal.pcbi.1002495
Simulation of molecular data under diverse evolutionary scenarios
Conflict of interest statement
The author has declared that no competing interests exist.
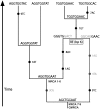
Figures

References
-
- Peck SL. Simulation as experiment: a philosophical reassessment for biological modeling. Trends Ecol Evol. 2004;19:530–534. - PubMed
-
- DeChaine EG, Martin AP. Using coalescent simulations to test the impact of quaternary climate cycles on divergence in an alpine plant-insect association. Evolution. 2006;60:1004–1013. - PubMed
-
- Westesson O, Holmes I. Accurate detection of recombinant breakpoints in whole-genome alignments. PLoS Comput Biol. 2009;5:e1000318. doi: 10.1371/journal.pcbi.1000318. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

