Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape
- PMID: 22693447
- PMCID: PMC3364956
- DOI: 10.1371/journal.ppat.1002721
Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape
Abstract
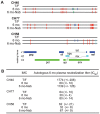
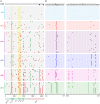
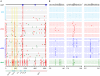
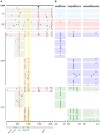
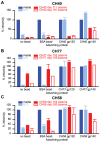
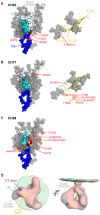
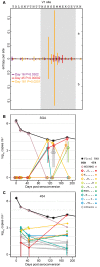
Single genome sequencing of early HIV-1 genomes provides a sensitive, dynamic assessment of virus evolution and insight into the earliest anti-viral immune responses in vivo. By using this approach, together with deep sequencing, site-directed mutagenesis, antibody adsorptions and virus-entry assays, we found evidence in three subjects of neutralizing antibody (Nab) responses as early as 2 weeks post-seroconversion, with Nab titers as low as 1∶20 to 1∶50 (IC(50)) selecting for virus escape. In each of the subjects, Nabs targeted different regions of the HIV-1 envelope (Env) in a strain-specific, conformationally sensitive manner. In subject CH40, virus escape was first mediated by mutations in the V1 region of the Env, followed by V3. HIV-1 specific monoclonal antibodies from this subject mapped to an immunodominant region at the base of V3 and exhibited neutralizing patterns indistinguishable from polyclonal antibody responses, indicating V1-V3 interactions within the Env trimer. In subject CH77, escape mutations mapped to the V2 region of Env, several of which selected for alterations of glycosylation. And in subject CH58, escape mutations mapped to the Env outer domain. In all three subjects, initial Nab recognition was followed by sequential rounds of virus escape and Nab elicitation, with Nab escape variants exhibiting variable costs to replication fitness. Although delayed in comparison with autologous CD8 T-cell responses, our findings show that Nabs appear earlier in HIV-1 infection than previously recognized, target diverse sites on HIV-1 Env, and impede virus replication at surprisingly low titers. The unexpected in vivo sensitivity of early transmitted/founder virus to Nabs raises the possibility that similarly low concentrations of vaccine-induced Nabs could impair virus acquisition in natural HIV-1 transmission, where the risk of infection is low and the number of viruses responsible for transmission and productive clinical infection is typically one.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. - PubMed
-
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. - PubMed
-
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. - PubMed
-
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI067854/AI/NIAID NIH HHS/United States
- R37 AI028433/AI/NIAID NIH HHS/United States
- AI67854/AI/NIAID NIH HHS/United States
- R01 RR006555/RR/NCRR NIH HHS/United States
- U01 AI041530/AI/NIAID NIH HHS/United States
- AI64518/AI/NIAID NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- P30 AI064518/AI/NIAID NIH HHS/United States
- AI27767/AI/NIAID NIH HHS/United States
- AI50410/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- R01 OD011095/OD/NIH HHS/United States
- AI61734/AI/NIAID NIH HHS/United States
- P01 AI061734/AI/NIAID NIH HHS/United States
- AI41530/AI/NIAID NIH HHS/United States
- AI028433/AI/NIAID NIH HHS/United States
- RR006555/RR/NCRR NIH HHS/United States
- R01 AI028433/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

