Allelic variation and differential expression of the mSIN3A histone deacetylase complex gene Arid4b promote mammary tumor growth and metastasis
- PMID: 22693453
- PMCID: PMC3364935
- DOI: 10.1371/journal.pgen.1002735
Allelic variation and differential expression of the mSIN3A histone deacetylase complex gene Arid4b promote mammary tumor growth and metastasis
Abstract
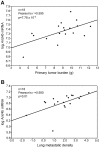
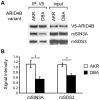
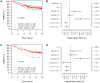
Accumulating evidence suggests that breast cancer metastatic progression is modified by germline polymorphism, although specific modifier genes have remained largely undefined. In the current study, we employ the MMTV-PyMT transgenic mouse model and the AKXD panel of recombinant inbred mice to identify AT-rich interactive domain 4B (Arid4b; NM_194262) as a breast cancer progression modifier gene. Ectopic expression of Arid4b promoted primary tumor growth in vivo as well as increased migration and invasion in vitro, and the phenotype was associated with polymorphisms identified between the AKR/J and DBA/2J alleles as predicted by our genetic analyses. Stable shRNA-mediated knockdown of Arid4b caused a significant reduction in pulmonary metastases, validating a role for Arid4b as a metastasis modifier gene. ARID4B physically interacts with the breast cancer metastasis suppressor BRMS1, and we detected differential binding of the Arid4b alleles to histone deacetylase complex members mSIN3A and mSDS3, suggesting that the mechanism of Arid4b action likely involves interactions with chromatin modifying complexes. Downregulation of the conserved Tpx2 gene network, which is comprised of many factors regulating cell cycle and mitotic spindle biology, was observed concomitant with loss of metastatic efficiency in Arid4b knockdown cells. Consistent with our genetic analysis and in vivo experiments in our mouse model system, ARID4B expression was also an independent predictor of distant metastasis-free survival in breast cancer patients with ER+ tumors. These studies support a causative role of ARID4B in metastatic progression of breast cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b.Cancer Res. 2013 Apr 15;73(8):2671-81. doi: 10.1158/0008-5472.CAN-12-3513. Epub 2013 Feb 27. Cancer Res. 2013. PMID: 23447578 Free PMC article.
-
Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells.J Biol Chem. 2008 Mar 21;283(12):7438-44. doi: 10.1074/jbc.M709446200. Epub 2008 Jan 22. J Biol Chem. 2008. PMID: 18211900 Free PMC article.
-
Ubiquitous Brms1 expression is critical for mammary carcinoma metastasis suppression via promotion of apoptosis.Clin Exp Metastasis. 2012 Apr;29(4):315-25. doi: 10.1007/s10585-012-9452-x. Epub 2012 Jan 13. Clin Exp Metastasis. 2012. PMID: 22241150 Free PMC article.
-
Metastasis suppression by BRMS1 associated with SIN3 chromatin remodeling complexes.Cancer Metastasis Rev. 2012 Dec;31(3-4):641-51. doi: 10.1007/s10555-012-9363-y. Cancer Metastasis Rev. 2012. PMID: 22678236 Review.
-
Breast cancer metastasis suppressor 1: update.Clin Exp Metastasis. 2003;20(1):45-50. doi: 10.1023/a:1022542519586. Clin Exp Metastasis. 2003. PMID: 12650606 Review.
Cited by
-
ceRNA Networks: The Backbone Role in Neoadjuvant Chemoradiotherapy Resistance/Sensitivity of Locally Advanced Rectal Cancer.Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211062313. doi: 10.1177/15330338211062313. Technol Cancer Res Treat. 2021. PMID: 34908512 Free PMC article.
-
Genome-wide patterns of genetic variation in two domestic chickens.Genome Biol Evol. 2013;5(7):1376-92. doi: 10.1093/gbe/evt097. Genome Biol Evol. 2013. PMID: 23814129 Free PMC article.
-
Expression and prognostic values of ARID family members in breast cancer.Aging (Albany NY). 2021 Feb 11;13(4):5621-5637. doi: 10.18632/aging.202489. Epub 2021 Feb 11. Aging (Albany NY). 2021. PMID: 33592583 Free PMC article.
-
Aicardi-Goutières syndrome gene Rnaseh2c is a metastasis susceptibility gene in breast cancer.PLoS Genet. 2019 May 24;15(5):e1008020. doi: 10.1371/journal.pgen.1008020. eCollection 2019 May. PLoS Genet. 2019. PMID: 31125342 Free PMC article.
-
CRISPR-Cas9 Library Screening Identifies Novel Molecular Vulnerabilities in KMT2A-Rearranged Acute Lymphoblastic Leukemia.Int J Mol Sci. 2023 Aug 25;24(17):13207. doi: 10.3390/ijms241713207. Int J Mol Sci. 2023. PMID: 37686014 Free PMC article.
References
-
- American Cancer Society. Cancer Facts and Figures 2012. Atlanta: American Cancer Society; 2012.
-
- Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–644. - PubMed
-
- Hunter KW, Broman KW, Voyer TL, Lukes L, Cozma D, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61:8866–8872. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

