Histone H3 localizes to the centromeric DNA in budding yeast
- PMID: 22693454
- PMCID: PMC3364953
- DOI: 10.1371/journal.pgen.1002739
Histone H3 localizes to the centromeric DNA in budding yeast
Abstract
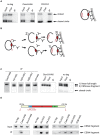
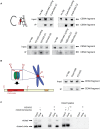
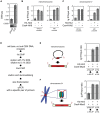
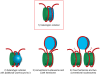
During cell division, segregation of sister chromatids to daughter cells is achieved by the poleward pulling force of microtubules, which attach to the chromatids by means of a multiprotein complex, the kinetochore. Kinetochores assemble at the centromeric DNA organized by specialized centromeric nucleosomes. In contrast to other eukaryotes, which typically have large repetitive centromeric regions, budding yeast CEN DNA is defined by a 125 bp sequence and assembles a single centromeric nucleosome. In budding yeast, as well as in other eukaryotes, the Cse4 histone variant (known in vertebrates as CENP-A) is believed to substitute for histone H3 at the centromeric nucleosome. However, the exact composition of the CEN nucleosome remains a subject of debate. We report the use of a novel ChIP approach to reveal the composition of the centromeric nucleosome and its localization on CEN DNA in budding yeast. Surprisingly, we observed a strong interaction of H3, as well as Cse4, H4, H2A, and H2B, but not histone chaperone Scm3 (HJURP in human) with the centromeric DNA. H3 localizes to centromeric DNA at all stages of the cell cycle. Using a sequential ChIP approach, we could demonstrate the co-occupancy of H3 and Cse4 at the CEN DNA. Our results favor a H3-Cse4 heterotypic octamer at the budding yeast centromere. Whether or not our model is correct, any future model will have to account for the stable association of histone H3 with the centromeric DNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. - PubMed
-
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

