The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway
- PMID: 22693585
- PMCID: PMC3365098
- DOI: 10.1371/journal.pone.0037925
The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway
Abstract
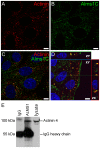
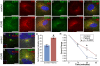
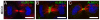
Alström syndrome (ALMS) is a progressive multi-systemic disorder characterized by cone-rod dystrophy, sensorineural hearing loss, childhood obesity, insulin resistance and cardiac, renal, and hepatic dysfunction. The gene responsible for Alström syndrome, ALMS1, is ubiquitously expressed and has multiple splice variants. The protein encoded by this gene has been implicated in ciliary function, cell cycle control, and intracellular transport. To gain better insight into the pathways through which ALMS1 functions, we carried out a yeast two hybrid (Y2H) screen in several mouse tissue libraries to identify ALMS1 interacting partners. The majority of proteins found to interact with the murine carboxy-terminal end (19/32) of ALMS1 were α-actinin isoforms. Interestingly, several of the identified ALMS1 interacting partners (α-actinin 1, α-actinin 4, myosin Vb, rad50 interacting 1 and huntingtin associated protein1A) have been previously associated with endosome recycling and/or centrosome function. We examined dermal fibroblasts from human subjects bearing a disruption in ALMS1 for defects in the endocytic pathway. Fibroblasts from these patients had a lower uptake of transferrin and reduced clearance of transferrin compared to controls. Antibodies directed against ALMS1 N- and C-terminal epitopes label centrosomes and endosomal structures at the cleavage furrow of dividing MDCK cells, respectively, suggesting isoform-specific cellular functions. Our results suggest a role for ALMS1 variants in the recycling endosome pathway and give us new insights into the pathogenesis of a subset of clinical phenotypes associated with ALMS.
Conflict of interest statement
Figures




References
-
- Marshall JD, Bronson RT, Collin GB, Nordstrom AD, Maffei P, et al. New Alstrom syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med. 2005;165:675–683. - PubMed
-
- Collin GB, Marshall JD, Ikeda A, So WV, Russell-Eggitt I, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31:74–78. - PubMed
-
- Hearn T, Renforth GL, Spalluto C, Hanley NA, Piper K, et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alstrom syndrome. Nat Genet. 2002;31:79–83. - PubMed
-
- Marshall JD, Hinman EG, Collin GB, Beck S, Cerqueira R, et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alstrom syndrome. Hum Mutat. 2007;28:1114–1123. - PubMed
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

