Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak
- PMID: 22693998
- PMCID: PMC3715836
- DOI: 10.1056/NEJMoa1109910
Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak
Abstract
Background: Isolates of methicillin-resistant Staphylococcus aureus (MRSA) belonging to a single lineage are often indistinguishable by means of current typing techniques. Whole-genome sequencing may provide improved resolution to define transmission pathways and characterize outbreaks.
Methods: We investigated a putative MRSA outbreak in a neonatal intensive care unit. By using rapid high-throughput sequencing technology with a clinically relevant turnaround time, we retrospectively sequenced the DNA from seven isolates associated with the outbreak and another seven MRSA isolates associated with carriage of MRSA or bacteremia in the same hospital.
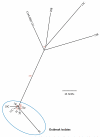
Results: We constructed a phylogenetic tree by comparing single-nucleotide polymorphisms (SNPs) in the core genome to a reference genome (an epidemic MRSA clone, EMRSA-15 [sequence type 22]). This revealed a distinct cluster of outbreak isolates and clear separation between these and the nonoutbreak isolates. A previously missed transmission event was detected between two patients with bacteremia who were not part of the outbreak. We created an artificial "resistome" of antibiotic-resistance genes and demonstrated concordance between it and the results of phenotypic susceptibility testing; we also created a "toxome" consisting of toxin genes. One outbreak isolate had a hypermutator phenotype with a higher number of SNPs than the other outbreak isolates, highlighting the difficulty of imposing a simple threshold for the number of SNPs between isolates to decide whether they are part of a recent transmission chain.
Conclusions: Whole-genome sequencing can provide clinically relevant data within a time frame that can influence patient care. The need for automated data interpretation and the provision of clinically meaningful reports represent hurdles to clinical implementation. (Funded by the U.K. Clinical Research Collaboration Translational Infection Research Initiative and others.).
Figures


References
-
- Baldry S. Attack of the clones. Nat Rev Microbiol. 2010;8:390. - PubMed
-
- Otto TD. Real-time sequencing. Nat Rev Microbiol. 2011;9:633. - PubMed
-
- Gardy JL, Johnston JC, Ho Sui SJ, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–9. Erratum, N Engl J Med 2011;364:2174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
