Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume
- PMID: 22695863
- PMCID: PMC3504958
- DOI: 10.1038/ismej.2012.64
Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume
Abstract
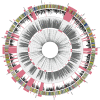
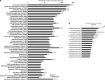
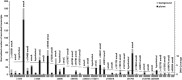
Ammonia-oxidizing Archaea (AOA) are among the most abundant microorganisms in the oceans and have crucial roles in biogeochemical cycling of nitrogen and carbon. To better understand AOA inhabiting the deep sea, we obtained community genomic and transcriptomic data from ammonium-rich hydrothermal plumes in the Guaymas Basin (GB) and from surrounding deep waters of the Gulf of California. Among the most abundant and active lineages in the sequence data were marine group I (MGI) Archaea related to the cultured autotrophic ammonia-oxidizer, Nitrosopumilus maritimus. Assembly of MGI genomic fragments yielded 2.9 Mb of sequence containing seven 16S rRNA genes (95.4-98.4% similar to N. maritimus), including two near-complete genomes and several lower-abundance variants. Equal copy numbers of MGI 16S rRNA genes and ammonia monooxygenase genes and transcription of ammonia oxidation genes indicates that all of these genotypes actively oxidize ammonia. De novo genomic assembly revealed the functional potential of MGI populations and enhanced interpretation of metatranscriptomic data. Physiological distinction from N. maritimus is evident in the transcription of novel genes, including genes for urea utilization, suggesting an alternative source of ammonia. We were also able to determine which genotypes are most active in the plume. Transcripts involved in nitrification were more prominent in the plume and were among the most abundant transcripts in the community. These unique data sets reveal populations of deep-sea AOA thriving in the ammonium-rich GB that are related to surface types, but with key genomic and physiological differences.
Figures


References
-
- Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–792. - PubMed
-
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. - PubMed
-
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

