Structural variants in the soybean genome localize to clusters of biotic stress-response genes
- PMID: 22696021
- PMCID: PMC3425179
- DOI: 10.1104/pp.112.194605
Structural variants in the soybean genome localize to clusters of biotic stress-response genes
Abstract
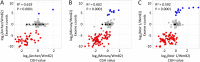
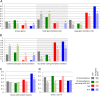
Genome-wide structural and gene content variations are hypothesized to drive important phenotypic variation within a species. Structural and gene content variations were assessed among four soybean (Glycine max) genotypes using array hybridization and targeted resequencing. Many chromosomes exhibited relatively low rates of structural variation (SV) among genotypes. However, several regions exhibited both copy number and presence-absence variation, the most prominent found on chromosomes 3, 6, 7, 16, and 18. Interestingly, the regions most enriched for SV were specifically localized to gene-rich regions that harbor clustered multigene families. The most abundant classes of gene families associated with these regions were the nucleotide-binding and receptor-like protein classes, both of which are important for plant biotic defense. The colocalization of SV with plant defense response signal transduction pathways provides insight into the mechanisms of soybean resistance gene evolution and may inform the development of new approaches to resistance gene cloning.
Figures



References
-
- Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, Peñuela S, Meksem K, Lightfoot DA, Young ND, et al. (2003) Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol Plant Microbe Interact 16: 817–826 - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

