Parallel MR imaging
- PMID: 22696125
- PMCID: PMC4459721
- DOI: 10.1002/jmri.23639
Parallel MR imaging
Abstract
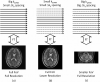
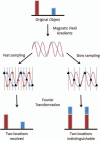
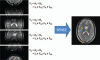
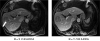
Parallel imaging is a robust method for accelerating the acquisition of magnetic resonance imaging (MRI) data, and has made possible many new applications of MR imaging. Parallel imaging works by acquiring a reduced amount of k-space data with an array of receiver coils. These undersampled data can be acquired more quickly, but the undersampling leads to aliased images. One of several parallel imaging algorithms can then be used to reconstruct artifact-free images from either the aliased images (SENSE-type reconstruction) or from the undersampled data (GRAPPA-type reconstruction). The advantages of parallel imaging in a clinical setting include faster image acquisition, which can be used, for instance, to shorten breath-hold times resulting in fewer motion-corrupted examinations. In this article the basic concepts behind parallel imaging are introduced. The relationship between undersampling and aliasing is discussed and two commonly used parallel imaging methods, SENSE and GRAPPA, are explained in detail. Examples of artifacts arising from parallel imaging are shown and ways to detect and mitigate these artifacts are described. Finally, several current applications of parallel imaging are presented and recent advancements and promising research in parallel imaging are briefly reviewed.
Copyright © 2012 Wiley Periodicals, Inc.
Figures










References
-
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. - PubMed
-
- Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47:1202–1210. - PubMed
-
- Paschal CB, Morris CB. K-space in the clinic. J Magn Reson Imaging. 2004;19:145–159. - PubMed
-
- Cohen MS, Weisskoff RM, Rzedzian RR, Kantor HL. Sensory stimulation by time-varying magnetic fields. Magn Reson Med. 1990;14:409–414. - PubMed
-
- Ham CL, Engels JM, van de Wiel GT, Machielsen A. Peripheral nerve stimulation during MRI: effects of high gradient amplitudes and switching rates. J Magn Reson Imaging. 1997;7:933–937. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

