The "Bridge" in the Epstein-Barr virus alkaline exonuclease protein BGLF5 contributes to shutoff activity during productive infection
- PMID: 22696660
- PMCID: PMC3416140
- DOI: 10.1128/JVI.00309-12
The "Bridge" in the Epstein-Barr virus alkaline exonuclease protein BGLF5 contributes to shutoff activity during productive infection
Abstract
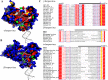
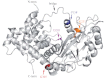
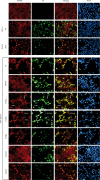
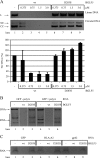
Replication of the human herpesvirus Epstein-Barr virus drastically impairs cellular protein synthesis. This shutoff phenotype results from mRNA degradation upon expression of the early lytic-phase protein BGLF5. Interestingly, BGLF5 is the viral DNase, or alkaline exonuclease, homologues of which are present throughout the herpesvirus family. During productive infection, this DNase is essential for processing and packaging of the viral genome. In contrast to this widely conserved DNase activity, shutoff is only mediated by the alkaline exonucleases of the subfamily of gammaherpesviruses. Here, we show that BGLF5 can degrade mRNAs of both cellular and viral origin, irrespective of polyadenylation. Furthermore, shutoff by BGLF5 induces nuclear relocalization of the cytosolic poly(A) binding protein. Guided by the recently resolved BGLF5 structure, mutants were generated and analyzed for functional consequences on DNase and shutoff activities. On the one hand, a point mutation destroying DNase activity also blocks RNase function, implying that both activities share a catalytic site. On the other hand, other mutations are more selective, having a more pronounced effect on either DNA degradation or shutoff. The latter results are indicative of an oligonucleotide-binding site that is partially shared by DNA and RNA. For this, the flexible "bridge" that crosses the active-site canyon of BGLF5 appears to contribute to the interaction with RNA substrates. These findings extend our understanding of the molecular basis for the shutoff function of BGLF5 that is conserved in gammaherpesviruses but not in alpha- and betaherpesviruses.
Figures



References
-
- Barnstable CJ, et al. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14:9–20 - PubMed
-
- Buisson M, et al. 2009. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 391:717–728 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

