Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway
- PMID: 22696683
- PMCID: PMC3408425
- DOI: 10.1091/mbc.E12-03-0232
Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway
Abstract
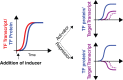
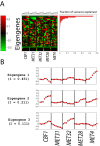
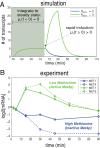
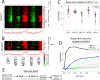
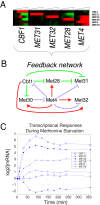
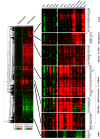
In yeast, the pathways of sulfur assimilation are combinatorially controlled by five transcriptional regulators (three DNA-binding proteins [Met31p, Met32p, and Cbf1p], an activator [Met4p], and a cofactor [Met28p]) and a ubiquitin ligase subunit (Met30p). This regulatory system exerts combinatorial control not only over sulfur assimilation and methionine biosynthesis, but also on many other physiological functions in the cell. Recently we characterized a gene induction system that, upon the addition of an inducer, results in near-immediate transcription of a gene of interest under physiological conditions. We used this to perturb levels of single transcription factors during steady-state growth in chemostats, which facilitated distinction of direct from indirect effects of individual factors dynamically through quantification of the subsequent changes in genome-wide patterns of gene expression. We were able to show directly that Cbf1p acts sometimes as a repressor and sometimes as an activator. We also found circumstances in which Met31p/Met32p function as repressors, as well as those in which they function as activators. We elucidated and numerically modeled feedback relationships among the regulators, notably feedforward regulation of Met32p (but not Met31p) by Met4p that generates dynamic differences in abundance that can account for the differences in function of these two proteins despite their identical binding sites.
Figures


References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

