QuaMeter: multivendor performance metrics for LC-MS/MS proteomics instrumentation
- PMID: 22697456
- PMCID: PMC3730131
- DOI: 10.1021/ac300629p
QuaMeter: multivendor performance metrics for LC-MS/MS proteomics instrumentation
Abstract
LC-MS/MS-based proteomics studies rely on stable analytical system performance that can be evaluated by objective criteria. The National Institute of Standards and Technology (NIST) introduced the MSQC software to compute diverse metrics from experimental LC-MS/MS data, enabling quality analysis and quality control (QA/QC) of proteomics instrumentation. In practice, however, several attributes of the MSQC software prevent its use for routine instrument monitoring. Here, we present QuaMeter, an open-source tool that improves MSQC in several aspects. QuaMeter can directly read raw data from instruments manufactured by different vendors. The software can work with a wide variety of peptide identification software for improved reliability and flexibility. Finally, QC metrics implemented in QuaMeter are rigorously defined and tested. The source code and binary versions of QuaMeter are available under Apache 2.0 License at http://fenchurch.mc.vanderbilt.edu.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
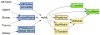
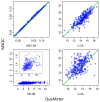
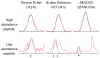

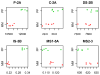
References
-
- Rudnick PA, Clauser KR, Kilpatrick LE, Tchekhovskoi DV, Neta P, Blonder N, Billheimer DD, Blackman RK, Bunk DM, Cardasis HL, Ham AJL, Jaffe JD, Kinsinger CR, Mesri M, Neubert TA, Schilling B, Tabb DL, Tegeler TJ, Vega-Montoto L, Variyath AM, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Carr SA, Fisher SJ, Gibson BW, Paulovich AG, Regnier FE, Rodriguez H, Spiegelman C, Tempst P, Liebler DC, Stein SE. Mol Cell Proteomics. 2010;9:225–241. - PMC - PubMed
-
- Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Proteomics. 2007;7:655–667. - PubMed
-
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J Proteome Res. 2004;3:958–964. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

