High codon adaptation in citrus tristeza virus to its citrus host
- PMID: 22698086
- PMCID: PMC3416656
- DOI: 10.1186/1743-422X-9-113
High codon adaptation in citrus tristeza virus to its citrus host
Abstract
Background: Citrus tristeza virus (CTV), a member of the genus Closterovirus within the family Closteroviridae, is the causal agent of citrus tristeza disease. Previous studies revealed that the negative selection, RNA recombination and gene flow were the most important forces that drove CTV evolution. However, the CTV codon usage was not studied and thus its role in CTV evolution remains unknown.
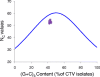
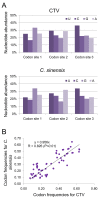
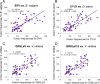
Results: A detailed comparative analysis of CTV codon usage pattern was done in this study. Results of the study show that although in general CTV does not have a high degree of codon usage bias, the codon usage of CTV has a high level of resemblance to its host codon usage. In addition, our data indicate that the codon usage resemblance is only observed for the woody plant-infecting closteroviruses but not the closteroviruses infecting the herbaceous host plants, suggesting the existence of different virus-host interactions between the herbaceous plant-infecting and woody plant-infecting closteroviruses.
Conclusion: Based on the results, we suggest that in addition to RNA recombination, negative selection and gene flow, host plant codon usage selection can also affect CTV evolution.
Figures

References
-
- Ermolaeva MD. Synonymous codon usage in bacteria. Curr Issues Mol Biol. 2001;3:91–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

