Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples
- PMID: 22698087
- PMCID: PMC3446314
- DOI: 10.1186/gb-2012-13-6-r42
Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples
Abstract
Background: To understand the relationship between our bacterial microbiome and health, it is essential to define the microbiome in the absence of disease. The digestive tract includes diverse habitats and hosts the human body's greatest bacterial density. We describe the bacterial community composition of ten digestive tract sites from more than 200 normal adults enrolled in the Human Microbiome Project, and metagenomically determined metabolic potentials of four representative sites.
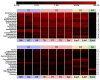
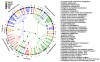
Results: The microbiota of these diverse habitats formed four groups based on similar community compositions: buccal mucosa, keratinized gingiva, hard palate; saliva, tongue, tonsils, throat; sub- and supra-gingival plaques; and stool. Phyla initially identified from environmental samples were detected throughout this population, primarily TM7, SR1, and Synergistetes. Genera with pathogenic members were well-represented among this disease-free cohort. Tooth-associated communities were distinct, but not entirely dissimilar, from other oral surfaces. The Porphyromonadaceae, Veillonellaceae and Lachnospiraceae families were common to all sites, but the distributions of their genera varied significantly. Most metabolic processes were distributed widely throughout the digestive tract microbiota, with variations in metagenomic abundance between body habitats. These included shifts in sugar transporter types between the supragingival plaque, other oral surfaces, and stool; hydrogen and hydrogen sulfide production were also differentially distributed.
Conclusions: The microbiomes of ten digestive tract sites separated into four types based on composition. A core set of metabolic pathways was present across these diverse digestive tract habitats. These data provide a critical baseline for future studies investigating local and systemic diseases affecting human health.
Figures




References
-
- Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, Rossmann P, Hrnčíř T, Kverka M, Zákostelská Z, Klimešová K, Přibylová J, Bártová J, Sanchez D, Fundová P, Borovská D, Srůtková D, Zídek Z, Schwarzer M, Drastich P, Funda DP. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. - DOI - PMC - PubMed
-
- Creely SJ, McTernan PG, Kusminski CM, Fisher M, Da Silva NF, Khanolkar M, Evans M, Harte AL, Kumar S. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292:E740–747. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

