The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B
- PMID: 22698963
- PMCID: PMC3422214
- DOI: 10.1016/j.abb.2012.06.002
The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B
Abstract
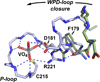
The movement of a conserved protein loop (the WPD-loop) is important in catalysis by protein tyrosine phosphatases (PTPs). Using kinetics, isotope effects, and X-ray crystallography, the different effects arising from mutation of the conserved tryptophan in the WPD-loop were compared in two PTPs, the human PTP1B, and the bacterial YopH from Yersinia. Mutation of the conserved tryptophan in the WPD-loop to phenylalanine has a negligible effect on k(cat) in PTP1B and full loop movement is maintained. In contrast, the corresponding mutation in YopH reduces k(cat) by two orders of magnitude and the WPD loop locks in an intermediate position, disabling general acid catalysis. During loop movement the indole moiety of the WPD-loop tryptophan moves in opposite directions in the two enzymes. Comparisons of mammalian and bacterial PTPs reveal differences in the residues forming the hydrophobic pocket surrounding the conserved tryptophan. Thus, although WPD-loop movement is a conserved feature in PTPs, differences exist in the molecular details, and in the tolerance to mutation, in PTP1B compared to YopH. Despite high structural similarity of the active sites in both WPD-loop open and closed conformations, differences are identified in the molecular details associated with loop movement in PTPs from different organisms.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
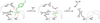
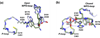

References
-
- Hunter T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signalling. Cell. 1995;80:225–236. - PubMed
-
- Jackson MD, Denu JM. Molecular reactions of protein phosphatases - Insights from structure and chemistry. Chemical Reviews. 2001;101:2313–2340. - PubMed
-
- Zhang Z-Y. Protein-tyrosine phosphatases: Biological function, structural characteristics, and mechanism of catalysis. Critical Reviews in Biochemistry and Molecular Biology. 1998;33:1–52. - PubMed
-
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

