High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance
- PMID: 22699502
- PMCID: PMC3396514
- DOI: 10.1073/pnas.1208003109
High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance
Erratum in
- Proc Natl Acad Sci U S A. 2012 Aug 7;109(32):13130
- Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):19969
Abstract
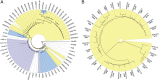
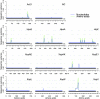
Cassava bacterial blight (CBB), incited by Xanthomonas axonopodis pv. manihotis (Xam), is the most important bacterial disease of cassava, a staple food source for millions of people in developing countries. Here we present a widely applicable strategy for elucidating the virulence components of a pathogen population. We report Illumina-based draft genomes for 65 Xam strains and deduce the phylogenetic relatedness of Xam across the areas where cassava is grown. Using an extensive database of effector proteins from animal and plant pathogens, we identify the effector repertoire for each sequenced strain and use a comparative sequence analysis to deduce the least polymorphic of the conserved effectors. These highly conserved effectors have been maintained over 11 countries, three continents, and 70 y of evolution and as such represent ideal targets for developing resistance strategies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

