Optimal tests for rare variant effects in sequencing association studies
- PMID: 22699862
- PMCID: PMC3440237
- DOI: 10.1093/biostatistics/kxs014
Optimal tests for rare variant effects in sequencing association studies
Abstract
With development of massively parallel sequencing technologies, there is a substantial need for developing powerful rare variant association tests. Common approaches include burden and non-burden tests. Burden tests assume all rare variants in the target region have effects on the phenotype in the same direction and of similar magnitude. The recently proposed sequence kernel association test (SKAT) (Wu, M. C., and others, 2011. Rare-variant association testing for sequencing data with the SKAT. The American Journal of Human Genetics 89, 82-93], an extension of the C-alpha test (Neale, B. M., and others, 2011. Testing for an unusual distribution of rare variants. PLoS Genetics 7, 161-165], provides a robust test that is particularly powerful in the presence of protective and deleterious variants and null variants, but is less powerful than burden tests when a large number of variants in a region are causal and in the same direction. As the underlying biological mechanisms are unknown in practice and vary from one gene to another across the genome, it is of substantial practical interest to develop a test that is optimal for both scenarios. In this paper, we propose a class of tests that include burden tests and SKAT as special cases, and derive an optimal test within this class that maximizes power. We show that this optimal test outperforms burden tests and SKAT in a wide range of scenarios. The results are illustrated using simulation studies and triglyceride data from the Dallas Heart Study. In addition, we have derived sample size/power calculation formula for SKAT with a new family of kernels to facilitate designing new sequence association studies.
Figures
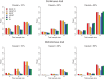
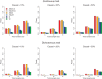

 versus t-statistic values of each variant of ANGPTL3, 4, and 5 genes. The dashed line represents the 95% confidence interval of no-association.
versus t-statistic values of each variant of ANGPTL3, 4, and 5 genes. The dashed line represents the 95% confidence interval of no-association.References
-
- Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., Hobbs H. H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869. - PubMed
-
- Davies R. B. Algorithm AS 155: the distribution of a linear combination of χ2 random variables. Applied Statistics. 1980;29:323–333.
-
- Davies R. B. Hypothesis testing when a nuisance parameter is present only under the alternative. Biometrika. 1987;74:33. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

