Research resource: Diagnostic and therapeutic potential of nuclear receptor expression in lung cancer
- PMID: 22700587
- PMCID: PMC3404298
- DOI: 10.1210/me.2011-1382
Research resource: Diagnostic and therapeutic potential of nuclear receptor expression in lung cancer
Abstract
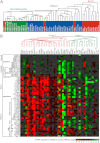
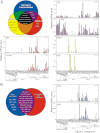
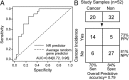
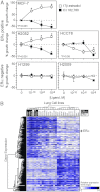
Lung cancer is the leading cause of cancer-related death. Despite a number of studies that have provided prognostic biomarkers for lung cancer, a paucity of reliable markers and therapeutic targets exist to diagnose and treat this aggressive disease. In this study we investigated the potential of nuclear receptors (NRs), many of which are well-established drug targets, as therapeutic markers in lung cancer. Using quantitative real-time PCR, we analyzed the expression of the 48 members of the NR superfamily in a human panel of 55 normal and lung cancer cell lines. Unsupervised cluster analysis of the NR expression profile segregated normal from tumor cell lines and grouped lung cancers according to type (i.e. small vs. non-small cell lung cancers). Moreover, we found that the NR signature was 79% accurate in diagnosing lung cancer incidence in smokers (n = 129). Finally, the evaluation of a subset of NRs (androgen receptor, estrogen receptor, vitamin D receptor, and peroxisome proliferator-activated receptor-γ) demonstrated the therapeutic potential of using NR expression to predict ligand-dependent growth responses in individual lung cancer cells. Preclinical evaluation of one of these receptors (peroxisome proliferator activated receptor-γ) in mouse xenografts confirmed that ligand-dependent inhibitory growth responses in lung cancer can be predicted based on a tumor's receptor expression status. Taken together, this study establishes NRs as theragnostic markers for predicting lung cancer incidence and further strengthens their potential as therapeutic targets for individualized treatment.
Figures



References
-
- Jemal A, Siegel R, Xu J, Ward E. 2010. Cancer statistics, 2010. CA Cancer J Clin 60:277–300 - PubMed
-
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. 2001. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 98:13790–13795 - PMC - PubMed
-
- Molina R, Augé JM, Bosch X, Escudero JM, Viñolas N, Marrades R, Ramírez J, Carcereny E, Filella X. 2009. Usefulness of serum tumor markers, including progastrin-releasing peptide, in patients with lung cancer: correlation with histology. Tumour Biol 30:121–129 - PubMed
-
- Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, Beer DG. 2006. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res 66:7466–7472 - PubMed
-
- Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, Sridhar S, Beamis J, Lamb C, Anderson T, Gerry N, Keane J, Lenburg ME, Brody JS. 2007. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 13:361–366 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

