Super-resolution imaging reveals the internal architecture of nano-sized syntaxin clusters
- PMID: 22700970
- PMCID: PMC3411058
- DOI: 10.1074/jbc.M112.353250
Super-resolution imaging reveals the internal architecture of nano-sized syntaxin clusters
Abstract
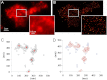
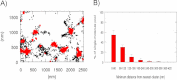
Key synaptic proteins from the soluble SNARE (N-ethylmaleimide-sensitive factor attachment protein receptor) family, among many others, are organized at the plasma membrane of cells as clusters containing dozens to hundreds of protein copies. However, the exact membranal distribution of proteins into clusters or as single molecules, the organization of molecules inside the clusters, and the clustering mechanisms are unclear due to limitations of the imaging and analytical tools. Focusing on syntaxin 1 and SNAP-25, we implemented direct stochastic optical reconstruction microscopy together with quantitative clustering algorithms to demonstrate a novel approach to explore the distribution of clustered and nonclustered molecules at the membrane of PC12 cells with single-molecule precision. Direct stochastic optical reconstruction microscopy images reveal, for the first time, solitary syntaxin/SNAP-25 molecules and small clusters as well as larger clusters. The nonclustered syntaxin or SNAP-25 molecules are mostly concentrated in areas adjacent to their own clusters. In the clusters, the density of the molecules gradually decreases from the dense cluster core to the periphery. We further detected large clusters that contain several density gradients. This suggests that some of the clusters are formed by unification of several clusters that preserve their original organization or reorganize into a single unit. Although syntaxin and SNAP-25 share some common distributional features, their clusters differ markedly from each other. SNAP-25 clusters are significantly larger, more elliptical, and less dense. Finally, this study establishes methodological tools for the analysis of single-molecule-based super-resolution imaging data and paves the way for revealing new levels of membranal protein organization.
Figures
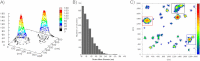



References
-
- Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 - PubMed
-
- Hanzal-Bayer M. F., Hancock J. F. (2007) Lipid rafts and membrane traffic. FEBS Lett. 581, 2098–2104 - PubMed
-
- Jacobson K., Mouritsen O. G., Anderson R. G. (2007) Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9, 7–14 - PubMed
-
- Shaw A. S. (2006) Lipid rafts: now you see them, now you don't. Nat. Immunol. 7, 1139–1142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

