Coordinated regulation of transcription factor Bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation
- PMID: 22700985
- PMCID: PMC3411033
- DOI: 10.1074/jbc.M112.344176
Coordinated regulation of transcription factor Bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation
Abstract
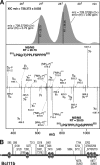
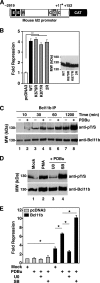
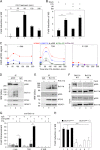
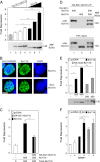
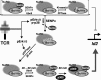
The transcriptional regulatory protein Bcl11b is essential for T-cell development. We have discovered a dynamic, MAPK-regulated pathway involving sequential, linked, and reversible post-translational modifications of Bcl11b in thymocytes. MAPK-mediated phosphorylation of Bcl11b was coupled to its rapid desumoylation, which was followed by a subsequent cycle of dephosphorylation and resumoylation. Additionally and notably, we report the first instance of direct identification by mass spectrometry of a site of small ubiquitin-like modifier (SUMO) adduction, Lys-679 of Bcl11b, in a protein isolated from a native, mammalian cell. Sumoylation of Bcl11b resulted in recruitment of the transcriptional co-activator p300 to a Bcl11b-repressed promoter with subsequent induction of transcription. Prolonged treatment of native thymocytes with phorbol 12,13-dibutyrate together with the calcium ionophore A23187 also promoted ubiquitination and proteasomal degradation of Bcl11b, providing a mechanism for signal termination. A Bcl11b phospho-deSUMO switch was identified, the basis of which was phosphorylation-dependent recruitment of the SUMO hydrolase SENP1 to phospho-Bcl11b, coupled to hydrolysis of SUMO-Bcl11b. These results define a regulatory pathway in thymocytes that includes the MAPK pathways and upstream signaling components, Bcl11b and the associated nucleosome remodeling and deacetylation (NuRD) complex, SENP proteins, the Bcl11b protein phosphatase 6, the sumoylation machinery, the histone acetyltransferase p300, and downstream transcriptional machinery. This pathway appears to facilitate derepression of repressed Bcl11b target genes as immature thymocytes initiate differentiation programs, biochemically linking MAPK signaling with the latter stages of T-cell development.
Figures





References
-
- Deribe Y. L., Pawson T., Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 - PubMed
-
- Johnson L. N., Lewis R. J. (2001) Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242 - PubMed
-
- Narayanan A., Jacobson M. P. (2009) Computational studies of protein regulation by post-translational phosphorylation. Curr. Opin. Struct. Biol. 19, 156–163 - PubMed
-
- Meulmeester E., Melchior F. (2008) Cell biology: SUMO. Nature 452, 709–711 - PubMed
-
- Tempé D., Piechaczyk M., Bossis G. (2008) SUMO under stress. Biochem. Soc. Trans. 36, 874–878 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

