A highly scalable peptide-based assay system for proteomics
- PMID: 22701568
- PMCID: PMC3373263
- DOI: 10.1371/journal.pone.0037441
A highly scalable peptide-based assay system for proteomics
Abstract
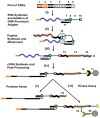
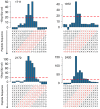
We report a scalable and cost-effective technology for generating and screening high-complexity customizable peptide sets. The peptides are made as peptide-cDNA fusions by in vitro transcription/translation from pools of DNA templates generated by microarray-based synthesis. This approach enables large custom sets of peptides to be designed in silico, manufactured cost-effectively in parallel, and assayed efficiently in a multiplexed fashion. The utility of our peptide-cDNA fusion pools was demonstrated in two activity-based assays designed to discover protease and kinase substrates. In the protease assay, cleaved peptide substrates were separated from uncleaved and identified by digital sequencing of their cognate cDNAs. We screened the 3,011 amino acid HCV proteome for susceptibility to cleavage by the HCV NS3/4A protease and identified all 3 known trans cleavage sites with high specificity. In the kinase assay, peptide substrates phosphorylated by tyrosine kinases were captured and identified by sequencing of their cDNAs. We screened a pool of 3,243 peptides against Abl kinase and showed that phosphorylation events detected were specific and consistent with the known substrate preferences of Abl kinase. Our approach is scalable and adaptable to other protein-based assays.
Conflict of interest statement
Figures




References
-
- Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program. National Human Genome Research Institute website. 2011;24 Available: www.genome.gov/sequencingcosts Accessed 2012 April.
-
- Scholle MD, Kriplani U, Pabon A, Sishtla K, Glucksman MJ, et al. Mapping protease substrates by using a biotinylated phage substrate library. Chembiochem. 2006;7:834–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

