Dramatic shifts in benthic microbial eukaryote communities following the Deepwater Horizon oil spill
- PMID: 22701662
- PMCID: PMC3368851
- DOI: 10.1371/journal.pone.0038550
Dramatic shifts in benthic microbial eukaryote communities following the Deepwater Horizon oil spill
Abstract
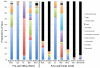
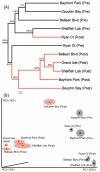
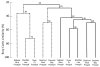
Benthic habitats harbour a significant (yet unexplored) diversity of microscopic eukaryote taxa, including metazoan phyla, protists, algae and fungi. These groups are thought to underpin ecosystem functioning across diverse marine environments. Coastal marine habitats in the Gulf of Mexico experienced visible, heavy impacts following the Deepwater Horizon oil spill in 2010, yet our scant knowledge of prior eukaryotic biodiversity has precluded a thorough assessment of this disturbance. Using a marker gene and morphological approach, we present an intensive evaluation of microbial eukaryote communities prior to and following oiling around heavily impacted shorelines. Our results show significant changes in community structure, with pre-spill assemblages of diverse Metazoa giving way to dominant fungal communities in post-spill sediments. Post-spill fungal taxa exhibit low richness and are characterized by an abundance of known hydrocarbon-degrading genera, compared to prior communities that contained smaller and more diverse fungal assemblages. Comparative taxonomic data from nematodes further suggests drastic impacts; while pre-spill samples exhibit high richness and evenness of genera, post-spill communities contain mainly predatory and scavenger taxa alongside an abundance of juveniles. Based on this community analysis, our data suggest considerable (hidden) initial impacts across Gulf beaches may be ongoing, despite the disappearance of visible surface oil in the region.
Conflict of interest statement
Figures
References
-
- Snelgrove PVR, Blackburn TH, Hutchings P, Alongi D, Grassle JF, et al. The importance of marine sediment biodiversity in ecosystem processes. Ambio. 1997;26:578–583.
-
- Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, et al. Ultrasequencing of the meiofaunal biosphere: practice, pitfalls, and promises. Molecular Ecology. 2010;19:4–20. - PubMed
-
- Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. The ISME Journal. 2008;2:997–1006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

