Differential gene expression segregates cattle confirmed positive for bovine tuberculosis from antemortem tuberculosis test-false positive cattle originating from herds free of bovine tuberculosis
- PMID: 22701814
- PMCID: PMC3373196
- DOI: 10.1155/2012/192926
Differential gene expression segregates cattle confirmed positive for bovine tuberculosis from antemortem tuberculosis test-false positive cattle originating from herds free of bovine tuberculosis
Abstract
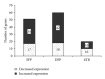
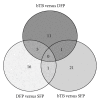
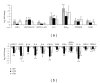
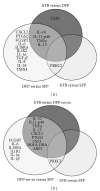
Antemortem tests for bovine tuberculosis (bTB) currently used in the US measure cell-mediated immune responses against Mycobacterium bovis. Postmortem tests for bTB rely on observation of gross and histologic lesions of bTB, followed by bacterial isolation or molecular diagnostics. Cumulative data from the state of Michigan indicates that 98 to 99% of cattle that react positively in antemortem tests are not confirmed positive for bTB at postmortem examination. Understanding the fundamental differences in gene regulation between antemortem test-false positive cattle and cattle that have bTB may allow identification of molecular markers that can be exploited to better separate infected from noninfected cattle. An immunospecific cDNA microarray was used to identify altered gene expression (P ≤ 0.01) of 122 gene features between antemortem test-false positive cattle and bTB-infected cattle following a 4-hour stimulation of whole blood with tuberculin. Further analysis using quantitative real-time PCR assays validated altered expression of 8 genes that had differential power (adj P ≤ 0.05) to segregate cattle confirmed positive for bovine tuberculosis from antemortem tuberculosis test-false positive cattle originating from herds free of bovine tuberculosis.
Figures
Similar articles
-
Use of whole-genome sequencing and evaluation of the apparent sensitivity and specificity of antemortem tuberculosis tests in the investigation of an unusual outbreak of Mycobacterium bovis infection in a Michigan dairy herd.J Am Vet Med Assoc. 2017 Jul 15;251(2):206-216. doi: 10.2460/javma.251.2.206. J Am Vet Med Assoc. 2017. PMID: 28671497
-
Antigen stimulation of peripheral blood mononuclear cells from Mycobacterium bovis infected cattle yields evidence for a novel gene expression program.BMC Genomics. 2008 Sep 29;9:447. doi: 10.1186/1471-2164-9-447. BMC Genomics. 2008. PMID: 18823559 Free PMC article.
-
Study on supplemental test to improve the detection of bovine tuberculosis in individual animals and herds.BMC Vet Res. 2021 Mar 31;17(1):137. doi: 10.1186/s12917-021-02839-4. BMC Vet Res. 2021. PMID: 33789652 Free PMC article.
-
Bovine TB and the development of new vaccines.Comp Immunol Microbiol Infect Dis. 2008 Mar;31(2-3):77-100. doi: 10.1016/j.cimid.2007.07.003. Epub 2007 Aug 30. Comp Immunol Microbiol Infect Dis. 2008. PMID: 17764740 Review.
-
Challenges for controlling bovine tuberculosis in South Africa.Onderstepoort J Vet Res. 2020 Feb 27;87(1):e1-e8. doi: 10.4102/ojvr.v87i1.1690. Onderstepoort J Vet Res. 2020. PMID: 32129639 Free PMC article. Review.
Cited by
-
Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals.Microorganisms. 2022 Sep 15;10(9):1845. doi: 10.3390/microorganisms10091845. Microorganisms. 2022. PMID: 36144447 Free PMC article. Review.
-
Ovine HSP90AA1 expression rate is affected by several SNPs at the promoter under both basal and heat stress conditions.PLoS One. 2013 Jun 24;8(6):e66641. doi: 10.1371/journal.pone.0066641. Print 2013. PLoS One. 2013. PMID: 23826107 Free PMC article.
References
-
- Schiller I, Oesch B, Vordermeier HM, et al. Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transboundary and Emerging Diseases. 2010;57(4):205–220. - PubMed
-
- O’Brien DJ, Schmitt SM, Fitzgerald SD, Berry DE, Hickling GJ. Managing the wildlife reservoir of Mycobacterium bovis: The Michigan, USA, experience. Veterinary Microbiology. 2006;112(2-4):313–323. - PubMed
-
- Michel AL, Bengis RG, Keet DF, et al. Wildlife tuberculosis in South African conservation areas: implications and challenges. Veterinary Microbiology. 2006;112(2-4):91–100. - PubMed
-
- de Kantor IN, Ambroggi M, Poggi S, et al. Human Mycobacterium bovis infection in ten Latin American countries. Tuberculosis. 2008;88(4):358–365. - PubMed
LinkOut - more resources
Full Text Sources

