Satellite cell heterogeneity revealed by G-Tool, an open algorithm to quantify myogenesis through colony-forming assays
- PMID: 22703589
- PMCID: PMC3439689
- DOI: 10.1186/2044-5040-2-13
Satellite cell heterogeneity revealed by G-Tool, an open algorithm to quantify myogenesis through colony-forming assays
Abstract
Background: Muscle growth and repair is accomplished by the satellite cell pool, a self-renewing population of myogenic progenitors. Functional heterogeneity within the satellite cell compartment and changes in potential with experimental intervention can be revealed by in vitro colony-forming cell (CFC) assays, however large numbers of colonies need to be assayed to give meaningful data, and manually quantifying nuclei and scoring markers of differentiation is experimentally limiting.
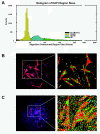
Methods: We present G-Tool, a multiplatform (Java) open-source algorithm that analyzes an ensemble of fluorescent micrographs of satellite cell-derived colonies to provide quantitative and statistically meaningful metrics of myogenic potential, including proliferation capacity and propensity to differentiate.
Results: We demonstrate the utility of G-Tool in two applications: first, we quantify the response of satellite cells to oxygen concentration. Compared to 3% oxygen which approximates tissue levels, we find that 21% oxygen, the ambient level, markedly limits the proliferative potential of transit amplifying progeny but at the same time inhibits the rate of terminal myogenic differentiation. We also test whether satellite cells from different muscles have intrinsic differences that can be read out in vitro. Compared to masseter, dorsi, forelimb and hindlimb muscles, we find that the diaphragm satellite cells have significantly increased proliferative potential and a reduced propensity to spontaneously differentiate. These features may be related to the unique always-active status of the diaphragm.
Conclusions: G-Tool facilitates consistent and reproducible CFC analysis between experiments and individuals. It is released under an open-source license that enables further development by interested members of the community.
Figures




References
-
- Mendell JR, Kissel JT, Amato AA, King W, Signore L, Prior TW, Sahenk Z, Benson S, McAndrew PE, Rice R, Nagaraja H, Stephens R, Lantry L, Morris GE, Burghes AHM. Myoblast transfer in the treatment of Duchenne’s muscular dystrophy. N Engl J Med. 1995;333:832–838. doi: 10.1056/NEJM199509283331303. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

