SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips
- PMID: 22703947
- PMCID: PMC3446316
- DOI: 10.1186/gb-2012-13-6-r44
SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips
Abstract
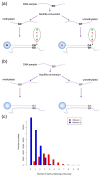
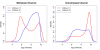
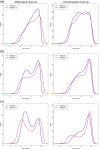
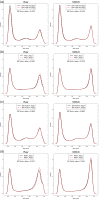
DNA methylation is the most widely studied epigenetic mark and is known to be essential to normal development and frequently disrupted in disease. The Illumina HumanMethylation450 BeadChip assays the methylation status of CpGs at 485,577 sites across the genome. Here we present Subset-quantile Within Array Normalization (SWAN), a new method that substantially improves the results from this platform by reducing technical variation within and between arrays. SWAN is available in the minfi Bioconductor package.
Figures


Similar articles
-
Complete pipeline for Infinium(®) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation.Epigenomics. 2012 Jun;4(3):325-41. doi: 10.2217/epi.12.21. Epigenomics. 2012. PMID: 22690668
-
Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi.Bioinformatics. 2017 Feb 15;33(4):558-560. doi: 10.1093/bioinformatics/btw691. Bioinformatics. 2017. PMID: 28035024 Free PMC article.
-
missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform.Bioinformatics. 2016 Jan 15;32(2):286-8. doi: 10.1093/bioinformatics/btv560. Epub 2015 Sep 30. Bioinformatics. 2016. PMID: 26424855
-
Analysis pipelines and packages for Infinium HumanMethylation450 BeadChip (450k) data.Methods. 2015 Jan 15;72:3-8. doi: 10.1016/j.ymeth.2014.08.011. Epub 2014 Sep 16. Methods. 2015. PMID: 25233806 Free PMC article. Review.
-
A comprehensive overview of Infinium HumanMethylation450 data processing.Brief Bioinform. 2014 Nov;15(6):929-41. doi: 10.1093/bib/bbt054. Epub 2013 Aug 29. Brief Bioinform. 2014. PMID: 23990268 Free PMC article. Review.
Cited by
-
Epigenetic dysregulation of immune-related pathways in cancer: bioinformatics tools and visualization.Exp Mol Med. 2021 May;53(5):761-771. doi: 10.1038/s12276-021-00612-z. Epub 2021 May 7. Exp Mol Med. 2021. PMID: 33963293 Free PMC article. Review.
-
Telomere length as a predictor of therapy response and survival in patients diagnosed with ovarian carcinoma.Heliyon. 2024 Jun 29;10(13):e33525. doi: 10.1016/j.heliyon.2024.e33525. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39050459 Free PMC article.
-
COHCAP: an integrative genomic pipeline for single-nucleotide resolution DNA methylation analysis.Nucleic Acids Res. 2013 Jun;41(11):e117. doi: 10.1093/nar/gkt242. Epub 2013 Apr 17. Nucleic Acids Res. 2013. PMID: 23598999 Free PMC article.
-
A systematic evaluation of normalization methods and probe replicability using infinium EPIC methylation data.Clin Epigenetics. 2023 Mar 11;15(1):41. doi: 10.1186/s13148-023-01459-z. Clin Epigenetics. 2023. PMID: 36906598 Free PMC article.
-
Epigenetic mechanism in search for the pathomechanism of diabetic neuropathy development in diabetes mellitus type 1 (T1DM).Endocrine. 2020 Apr;68(1):235-240. doi: 10.1007/s12020-019-02172-9. Epub 2020 Jan 4. Endocrine. 2020. PMID: 31902112
References
-
- Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, Tomazou EM, Bäckdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJP, Carter NP, Tavaré S, Beck S. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. - DOI - PMC - PubMed
-
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

