Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy
- PMID: 22704557
- PMCID: PMC3595537
- DOI: 10.1016/j.molcel.2012.05.027
Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy
Abstract
Macroautophagy is essential to cell survival during starvation and proceeds by the growth of a double-membraned phagophore, which engulfs cytosol and other substrates. The synthesis and recognition of the lipid phosphatidylinositol 3-phosphate, PI(3)P, is essential for autophagy. The key autophagic PI(3)P sensors, which are conserved from yeast to humans, belong to the PROPPIN family. Here we report the crystal structure of the yeast PROPPIN Hsv2. The structure consists of a seven-bladed β-propeller and, unexpectedly, contains two pseudo-equivalent PI(3)P binding sites on blades 5 and 6. These two sites both contribute to membrane binding in vitro and are collectively required for full autophagic function in yeast. These sites function in concert with membrane binding by a hydrophobic loop in blade 6, explaining the specificity of the PROPPINs for membrane-bound PI(3)P. These observations thus provide a structural and mechanistic framework for one of the conserved central molecular recognition events in autophagy.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

FRET assay for Hsv2 binding to PI(3)P, PI(3,5)P2 and control PS liposomes. Error bars represent the standard deviation of triplicate measurements.
Ribbon model of the Hsv2 structure, colored from blade 1–7 and the sulfates shown as spheres.
Surface representation of Hsv2 showing the discoidal shape and location of bound sulfate ions.
The conserved arginines of the FRRG motif coordinating the sulfates that are 17 Å apart in the two sites are shown in ball and stick model.
Ribbon representation of Hsv2 with the sulfates and binding site 1 (E) and 2 (F) residues in ball and stick model.

Structurally annotated alignment of PROPPIN sequences for lipid-binding blades 5 and 6. Sequences were aligned with T-Coffee within each blade based on multiple alignment of primary sequences. Residues showing 100, 80 and 60% conservation across the different proteins are highlighted in red, blue and green respectively.
Ribbon representation of the structural superposition of blades 5 (green) and 6 (pink). The sulfates and binding sites residues are shown in ball and stick model.
Mutations in the indicated site 1 and site 2 residues reduce or eliminate binding to liposomes.

Surface of Hsv2 colored according to electrostatic potential, with saturating blue and red at ±3 kT/e.
Charge reversal mutations (blue boxes and type) in the flat basic face of the disk do not perturb binding to liposomes, but hydrophobic loop mutations (green boxes and type) eliminate binding.
Docking of Hsv2 to a model PI(3)P-containing membrane.

Structurally annotated alignment of PROPPIN sequences for β3-β4 region in blade 6. The hydrophobic residues in the β3-β4 loop are highlighted in yellow.
Human WIPI4 which has one hydrophobic residue in the loop region binds more weakly to PI(3)P-containing liposomes than other PROPPINs.


atg18Δ S. cerevisiae cells transformed with ATG18-GFP, ATG18Site1-GFP, ATG18Site2-GFP, ATG18Site1/Site2-GFP and ATG18Site1/Site2/Δ3Ar-GFP were grown to mid log phase and visualized by confocal microscopy. Representative cells are shown with Atg18-GFP fluorescence images on the left and transmitted light images on the right.
Western blot analysis was performed using anti-GFP antibodies to determine the overall expression of each Atg18-GFP variant.
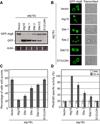
GFP-Atg8 processing monitored by western blot against GFP. The full length GFP-Atg8 and GFP bands are labeled. The actin loading control is shown below.
Representative microscopy images for GFP-Atg8 shown on the left and transmitted light images shown on the right.
The percentage of cells displaying GFP-Atg8 puncta with a total of 100 cells counted.
Pho8Δ60 alkaline phosphatase specific activity normalized to starved cells expressing Atg18-myc. Data for non-starved cells are shown in white and starved cells are shown in gray. Error bars in (C) and (D) represent the standard deviation of triplicate experiments.
References
-
- Barth H, Meiling-Wesse K, Epple UD, Thumm M. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 2001;508:23–28. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

