Selective and specific inhibition of the plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin
- PMID: 22704625
- PMCID: PMC3391680
- DOI: 10.1016/j.chom.2012.04.015
Selective and specific inhibition of the plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin
Abstract
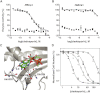
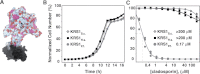
With renewed calls for malaria eradication, next-generation antimalarials need be active against drug-resistant parasites and efficacious against both liver- and blood-stage infections. We screened a natural product library to identify inhibitors of Plasmodium falciparum blood- and liver-stage proliferation. Cladosporin, a fungal secondary metabolite whose target and mechanism of action are not known for any species, was identified as having potent, nanomolar, antiparasitic activity against both blood and liver stages. Using postgenomic methods, including a yeast deletion strains collection, we show that cladosporin specifically inhibits protein synthesis by directly targeting P. falciparum cytosolic lysyl-tRNA synthetase. Further, cladosporin is >100-fold more potent against parasite lysyl-tRNA synthetase relative to the human enzyme, which is conferred by the identity of two amino acids within the enzyme active site. Our data indicate that lysyl-tRNA synthetase is an attractive, druggable, antimalarial target that can be selectively inhibited.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
An all-purpose antimalarial drug target.Cell Host Microbe. 2012 Jun 14;11(6):555-7. doi: 10.1016/j.chom.2012.05.008. Cell Host Microbe. 2012. PMID: 22704614 Review.
-
The growing pipeline of natural aminoacyl-tRNA synthetase inhibitors for malaria treatment.Bioengineered. 2016 Apr 2;7(2):60-4. doi: 10.1080/21655979.2016.1149270. Epub 2016 Mar 10. Bioengineered. 2016. PMID: 26963157 Free PMC article.
Similar articles
-
An all-purpose antimalarial drug target.Cell Host Microbe. 2012 Jun 14;11(6):555-7. doi: 10.1016/j.chom.2012.05.008. Cell Host Microbe. 2012. PMID: 22704614 Review.
-
Design and Synthesis of Metabolically Stable tRNA Synthetase Inhibitors Derived from Cladosporin.Chembiochem. 2019 Mar 1;20(5):644-649. doi: 10.1002/cbic.201800587. Epub 2019 Feb 4. Chembiochem. 2019. PMID: 30462880
-
Inhibition of Plasmodium falciparum Lysyl-tRNA Synthetase via a Piperidine-Ring Scaffold Inspired Cladosporin Analogues.Chembiochem. 2021 Jul 15;22(14):2468-2477. doi: 10.1002/cbic.202100212. Epub 2021 May 28. Chembiochem. 2021. PMID: 33969584
-
Cladosporin, A Highly Potent Antimalaria Drug?Chembiochem. 2023 Jun 15;24(12):e202300154. doi: 10.1002/cbic.202300154. Epub 2023 May 23. Chembiochem. 2023. PMID: 37158666 Review.
-
Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.J Struct Funct Genomics. 2014 Jun;15(2):63-71. doi: 10.1007/s10969-014-9182-1. Epub 2014 Jun 17. J Struct Funct Genomics. 2014. PMID: 24935905
Cited by
-
Genetic crosses within and between species of Cryptosporidium.Proc Natl Acad Sci U S A. 2024 Jan 2;121(1):e2313210120. doi: 10.1073/pnas.2313210120. Epub 2023 Dec 26. Proc Natl Acad Sci U S A. 2024. PMID: 38147547 Free PMC article.
-
Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development.Mol Microbiol. 2013 Aug;89(4):660-75. doi: 10.1111/mmi.12303. Epub 2013 Jul 12. Mol Microbiol. 2013. PMID: 23796209 Free PMC article.
-
A nondiscriminating glutamyl-tRNA synthetase in the plasmodium apicoplast: the first enzyme in an indirect aminoacylation pathway.J Biol Chem. 2013 Nov 8;288(45):32539-32552. doi: 10.1074/jbc.M113.507467. Epub 2013 Sep 26. J Biol Chem. 2013. PMID: 24072705 Free PMC article.
-
Chemical interrogation of the malaria kinome.Chembiochem. 2014 Sep 5;15(13):1920-30. doi: 10.1002/cbic.201400025. Epub 2014 Aug 8. Chembiochem. 2014. PMID: 25111632 Free PMC article.
-
The yeast deletion collection: a decade of functional genomics.Genetics. 2014 Jun;197(2):451-65. doi: 10.1534/genetics.114.161620. Epub 2014 Jun 17. Genetics. 2014. PMID: 24939991 Free PMC article. Review.
References
-
- Anke H. Metabolic products of microorganisms. 184. On the mode of action of cladosporin. J. Antibiot. (Tokyo) 1979;32:952–958. - PubMed
-
- Burrows J.N., Chibale K., Wells T.N. The state of the art in anti-malarial drug discovery and development. Curr. Top. Med. Chem. 2011;11:1226–1254. - PubMed
-
- Dharia N.V., Sidhu A.B., Cassera M.B., Westenberger S.J., Bopp S.E., Eastman R.T., Plouffe D., Batalov S., Park D.J., Volkman S.K. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. - PMC - PubMed
-
- Gamo F.J., Sanz L.M., Vidal J., de Cozar C., Alvarez E., Lavandera J.L., Vanderwall D.E., Green D.V., Kumar V., Hasan S. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

