Biosynthesis of albomycin δ(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates
- PMID: 22704654
- PMCID: PMC3448783
- DOI: 10.1021/cb300173x
Biosynthesis of albomycin δ(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates
Abstract
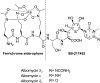
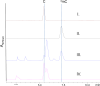
"Trojan horse" antibiotic albomycins are peptidyl nucleosides consisting of a highly modified 4'-thiofuranosyl cytosine moiety and a ferrichrome siderophore that are linked by a peptide bond via a serine residue. While the latter component serves to sequester iron from the environment, the seryl nucleoside portion is a potent inhibitor of bacterial seryl-tRNA synthetases, resulting in broad-spectrum antimicrobial activities of albomycin δ(2). The isolation of albomycins has revealed this biological activity is optimized only following two unusual cytosine modifications, N4-carbamoylation and N3-methylation. We identified a genetic locus (named abm) for albomycin production in Streptomyces sp. ATCC 700974. Gene deletion and complementation experiments along with bioinformatic analysis suggested 18 genes are responsible for albomycin biosynthesis and resistance, allowing us to propose a potential biosynthetic pathway for installing the novel chemical features. The gene abmI, encoding a putative methyltransferase, was functionally assigned in vitro and shown to modify the N3 of a variety of cytosine-containing nucleosides and antibiotics such as blasticidin S. Furthermore, a ΔabmI mutant was shown to produce the descarbamoyl-desmethyl albomycin analogue, supporting that the N3-methylation occurs before the N4-carbamoylation in the biosynthesis of albomycin δ(2). The combined genetic information was utilized to identify an abm-related locus (named ctj) from the draft genome of Streptomyces sp. C. Cross-complementation experiments and in vitro studies with CtjF, the AbmI homologue, suggest the production of a similar 4'-thiofuranosyl cytosine in this organism. In total, the genetic and biochemical data provide a biosynthetic template for assembling siderophore-inhibitor conjugates and modifying the albomycin scaffold to generate new derivatives.
Figures




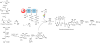
References
-
- Braznikova G. Iron-containing antibiotic produced by Actinomyces subtropicus. Nov. Med. 1951;23:3.
-
- Maehr H, Berger J. The production, isolation and characterization of a grisein-like sideromycin complex. Biotechnology and Bioengineering XI. 1969:1111–1123. - PubMed
-
- Garrod LP. Correspondence. British Medical Journal. 1956:1362.
-
- Paulsen H, Brieden M, Benz G. Synthese des Sauerstofanalogs der Desferriforn von δ1-Albomycine. Liebigs Ann Chem. 1987:565–575.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

