Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB
- PMID: 22705207
- PMCID: PMC3392440
- DOI: 10.1016/j.str.2012.04.021
Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB
Abstract
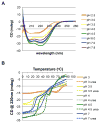
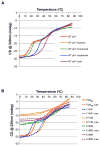
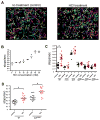
pH sensing is crucial for survival of most organisms, yet the molecular basis of such sensing is poorly understood. Here, we present an atomic resolution structure of the periplasmic portion of the acid-sensing chemoreceptor, TlpB, from the gastric pathogen Helicobacter pylori. The structure reveals a universal signaling fold, a PAS domain, with a molecule of urea bound with high affinity. Through biophysical, biochemical, and in vivo mutagenesis studies, we show that urea and the urea-binding site residues play critical roles in the ability of H. pylori to sense acid. Our signaling model predicts that protonation events at Asp114, affected by changes in pH, dictate the stability of TlpB through urea binding.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



References
-
- Bowie JU, Pakula AA, Simon MI. The three-dimensional structure of the aspartate receptor from Escherichia coli. Acta Crystallogr D Biol Crystallogr. 1995;51:145–154. - PubMed
-
- Bowie JU, Sauer RT. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989;28:7139–7143. - PubMed
-
- Celej MS, Dassie SA, Freire E, Bianconi ML, Fidelio GD. Ligand-induced thermostability in proteins: thermodynamic analysis of ANS-albumin interaction. Biochim Biophys Acta. 2005a;1750:122–133. - PubMed
-
- Celej MS, Fidelio GD, Dassie SA. Protein Unfolding Coupled to Ligand Binding: Differential Scanning Calorimetry Simulation Approach. J Chem Ed. 2005b;82:87–92.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

