Silent substitutions predictably alter translation elongation rates and protein folding efficiencies
- PMID: 22705285
- PMCID: PMC3576719
- DOI: 10.1016/j.jmb.2012.06.010
Silent substitutions predictably alter translation elongation rates and protein folding efficiencies
Abstract
Genetic code redundancy allows most amino acids to be encoded by multiple codons that are non-randomly distributed along coding sequences. An accepted theory explaining the biological significance of such non-uniform codon selection is that codons are translated at different speeds. Thus, varying codon placement along a message may confer variable rates of polypeptide emergence from the ribosome, which may influence the capacity to fold toward the native state. Previous studies report conflicting results regarding whether certain codons correlate with particular structural or folding properties of the encoded protein. This is partly due to different criteria traditionally utilized for predicting translation speeds of codons, including their usage frequencies and the concentration of tRNA species capable of decoding them, which do not always correlate. Here, we developed a metric to predict organism-specific relative translation rates of codons based on the availability of tRNA decoding mechanisms: Watson-Crick, non-Watson-Crick or both types of interactions. We determine translation rates of messages by pulse-chase analyses in living Escherichia coli cells and show that sequence engineering based on these concepts predictably modulates translation rates in a manner that is superior to codon usage frequency, which occur during the elongation phase, and significantly impacts folding of the encoded polypeptide. Finally, we demonstrate that sequence harmonization based on expression host tRNA pools, designed to mimic ribosome movement of the original organism, can significantly increase the folding of the encoded polypeptide. These results illuminate how genetic code degeneracy may function to specify properties beyond amino acid encoding, including folding.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

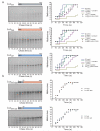

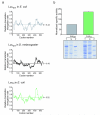
Comment in
-
Ribosomal kinetics and accuracy: sequence engineering to the rescue.J Mol Biol. 2012 Sep 21;422(3):325-7. doi: 10.1016/j.jmb.2012.07.002. Epub 2012 Jul 4. J Mol Biol. 2012. PMID: 22771360 No abstract available.
References
-
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. - PubMed
-
- Deane CM, Saunders R. The imprint of codons on protein structure. Biotechnol J. 2011;6:641–9. - PubMed
-
- Spencer PS, Barral JM. Genetic code redundancy and its influence on the encoded polypeptides. Computational and Structural Biotechnology Journal. 2012;1:e201204006. doi: http://dx.doi.org/10.5936/csbj.201204006. - DOI - PMC - PubMed
-
- Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell. 2008;30:589–98. - PubMed
-
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

