The brace for a growing scaffold: Mss116 protein promotes RNA folding by stabilizing an early assembly intermediate
- PMID: 22705286
- PMCID: PMC4419380
- DOI: 10.1016/j.jmb.2012.05.037
The brace for a growing scaffold: Mss116 protein promotes RNA folding by stabilizing an early assembly intermediate
Abstract
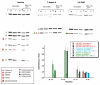
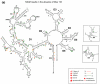
The ai5γ group II intron requires a protein cofactor to facilitate native folding in the cell. Yeast protein Mss116 greatly accelerates intron folding under near-physiological conditions both in vivo and in vitro. Although the effect of Mss116 on the kinetics of ai5γ ribozyme folding and catalysis has been extensively studied, the precise structural role and interaction sites of Mss116 have been elusive. Using Nucleotide Analog Interference Mapping to study the folding of splicing precursor constructs, we have identified specific intron functional groups that participate in Mss116-facilitated folding and we have determined their role in the folding mechanism. The data indicate that Mss116 stabilizes an early, obligate folding intermediate within intron domain 1, thereby laying the foundation for productive folding to the native state. In addition, the data reveal an important role for the IBS2 exon sequence and for the terminus of domain 6, during the folding of self-splicing group IIB intron constructs.
Copyright © 2012. Published by Elsevier Ltd.
Figures










References
-
- Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol. 2005;353:1199–11209. - PubMed
-
- Su L, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 2003;334:639–652. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

