Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation
- PMID: 22705549
- PMCID: PMC3399031
- DOI: 10.1016/j.bbrc.2012.06.033
Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation
Abstract
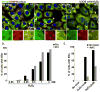
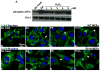
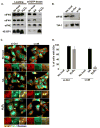
In cells exposed to environmental stress, inhibition of translation initiation conserves energy for the repair of cellular damage. Untranslated mRNAs that accumulate in these cells move to discrete cytoplasmic foci known as stress granules (SGs). The assembly of SGs helps cells to survive under adverse environmental conditions. We have analyzed the mechanism by which hydrogen peroxide (H(2)O(2))-induced oxidative stress inhibits translation initiation and induces SG assembly in mammalian cells. Our data indicate that H(2)O(2) inhibits translation and induces the assembly of SGs. The assembly of H(2)O(2)-induced SGs is independent of the phosphorylation of eIF2α, a major trigger of SG assembly, but requires remodeling of the cap-binding eIF4F complex. Moreover, H(2)O(2)-induced SGs are compositionally distinct from canonical SGs, and targeted knockdown of eIF4E, a protein required for canonical translation initiation, inhibits H(2)O(2)-induced SG assembly. Our data reveal new aspects of translational regulation induced by oxidative insults.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

