Identification and characterization of Cronobacter iron acquisition systems
- PMID: 22706064
- PMCID: PMC3416605
- DOI: 10.1128/AEM.01457-12
Identification and characterization of Cronobacter iron acquisition systems
Abstract
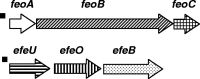
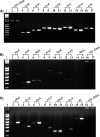
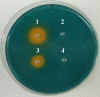
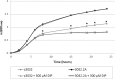
Cronobacter spp. are emerging pathogens that cause severe infantile meningitis, septicemia, or necrotizing enterocolitis. Contaminated powdered infant formula has been implicated as the source of Cronobacter spp. in most cases, but questions still remain regarding the natural habitat and virulence potential for each strain. The iron acquisition systems in 231 Cronobacter strains isolated from different sources were identified and characterized. All Cronobacter spp. have both the Feo and Efe systems for acquisition of ferrous iron, and all plasmid-harboring strains (98%) have the aerobactin-like siderophore, cronobactin, for transport of ferric iron. All Cronobacter spp. have the genes encoding an enterobactin-like siderophore, although it was not functional under the conditions tested. Furthermore, all Cronobacter spp. have genes encoding five receptors for heterologous siderophores. A ferric dicitrate transport system (fec system) is encoded specifically by a subset of Cronobacter sakazakii and C. malonaticus strains, of which a high percentage were isolated from clinical samples. Phylogenetic analysis confirmed that the fec system is most closely related to orthologous genes present in human-pathogenic bacterial strains. Moreover, all strains of C. dublinensis and C. muytjensii encode two receptors, FcuA and Fct, for heterologous siderophores produced by plant pathogens. Identification of putative Fur boxes and expression of the genes under iron-depleted conditions revealed which genes and operons are components of the Fur regulon. Taken together, these results support the proposition that C. sakazakii and C. malonaticus may be more associated with the human host and C. dublinensis and C. muytjensii with plants.
Figures



Similar articles
-
Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp.Appl Environ Microbiol. 2011 May;77(10):3255-67. doi: 10.1128/AEM.03023-10. Epub 2011 Mar 18. Appl Environ Microbiol. 2011. PMID: 21421789 Free PMC article.
-
Characterization of Cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic.Int J Food Microbiol. 2016 Jan 18;217:130-6. doi: 10.1016/j.ijfoodmicro.2015.10.017. Epub 2015 Oct 17. Int J Food Microbiol. 2016. PMID: 26513253
-
Genotyping and Source Tracking of Cronobacter sakazakii and C. malonaticus Isolates from Powdered Infant Formula and an Infant Formula Production Factory in China.Appl Environ Microbiol. 2015 Aug 15;81(16):5430-9. doi: 10.1128/AEM.01390-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048942 Free PMC article.
-
Cronobacter spp.--opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits.J Med Microbiol. 2014 Aug;63(Pt 8):1023-1037. doi: 10.1099/jmm.0.073742-0. Epub 2014 May 30. J Med Microbiol. 2014. PMID: 24878566 Review.
-
Strategies for the identification and tracking of cronobacter species: an opportunistic pathogen of concern to neonatal health.Front Pediatr. 2015 May 5;3:38. doi: 10.3389/fped.2015.00038. eCollection 2015. Front Pediatr. 2015. PMID: 26000266 Free PMC article. Review.
Cited by
-
Alterations in the Transcriptional Landscape Allow Differential Desiccation Tolerance in Clinical Cronobacter sakazakii.Appl Environ Microbiol. 2021 Nov 24;87(24):e0083021. doi: 10.1128/AEM.00830-21. Epub 2021 Oct 13. Appl Environ Microbiol. 2021. PMID: 34644165 Free PMC article.
-
Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria.3 Biotech. 2017 Dec;7(6):381. doi: 10.1007/s13205-017-1008-y. Epub 2017 Oct 26. 3 Biotech. 2017. PMID: 29109926 Free PMC article.
-
Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence.Front Cell Infect Microbiol. 2013 Dec 5;3:90. doi: 10.3389/fcimb.2013.00090. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24367764 Free PMC article. Review.
-
Cronobacter: an emergent pathogen causing meningitis to neonates through their feeds.Sci Prog. 2014;97(Pt 2):154-72. doi: 10.3184/003685014X13994743930498. Sci Prog. 2014. PMID: 25108996 Free PMC article.
-
Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility.Front Microbiol. 2013 Sep 2;4:256. doi: 10.3389/fmicb.2013.00256. eCollection 2013. Front Microbiol. 2013. PMID: 24032028 Free PMC article.
References
-
- Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11: 36–42
-
- Angerer A, Braun V. 1998. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 169: 483–490 - PubMed
-
- Arnow LE. 1937. Colorimetric determination of the components of 3,4-dihydroxy-phenylalanine-tyrosine mixture. J. Biol. Chem. 118: 531–541
-
- Bäumler AJ, Hantke K. 1992. Ferrioxamine uptake in Yersinia enterocolitica: characterization of the receptor protein FoxA. Mol. Microbiol. 6: 1309–1321 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

