Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells
- PMID: 22706276
- PMCID: PMC3428153
- DOI: 10.1093/hmg/dds224
Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells
Abstract
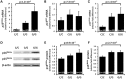
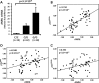
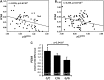
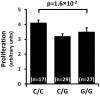
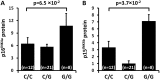
Variation on chromosome 9p21 is associated with risk of coronary artery disease (CAD). This genomic region contains the CDKN2A and CDKN2B genes which encode the cell cycle regulators p16(INK4a), p14(ARF) and p15(INK4b) and the ANRIL gene which encodes a non-coding RNA. Vascular smooth muscle cell (VSMC) proliferation plays an important role in the pathogenesis of atherosclerosis which causes CAD. We ascertained whether 9p21 genotype had an influence on CDKN2A/CDKN2B/ANRIL expression levels in VSMCs, VSMC proliferation and VSMC content in atherosclerotic plaques. Immunohistochemical examination showed that VSMCs in atherosclerotic lesions expressed p16(INK4a), p14(ARF) and p15(INK4b). Analyses of primary cultures of VSMCs showed that the 9p21 risk genotype was associated with reduced expression of p16(INK4a), p15(INK4b) and ANRIL (P = 1.2 × 10(-5), 1.4 × 10(-2) and 3.1 × 10(-9)) and with increased VSMC proliferation (P = 1.6 × 10(-2)). Immunohistochemical analyses of atherosclerotic plaques revealed an association of the risk genotype with reduced p15(INK4b) levels in VSMCs (P = 3.7 × 10(-2)) and higher VSMC content (P = 5.6 × 10(-4)) in plaques. The results of this study indicate that the 9p21 variation has an impact on CDKN2A and CDKN2B expression in VSMCs and influences VMSC proliferation, which likely represents an important mechanism for the association between this genetic locus and susceptibility to CAD.
Figures


References
-
- Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., et al. Genomewide association analysis of coronary artery disease. N. Engl J. Med. 2007;357:443–453. doi:10.1056/NEJMoa072366. - DOI - PMC - PubMed
-
- Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi:10.1126/science.1142842. - DOI - PubMed
-
- McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi:10.1126/science.1142447. - DOI - PMC - PubMed
-
- Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., Schunkert H., et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi:10.1038/ng.327. - DOI - PMC - PubMed
-
- Broadbent H.M., Peden J.F., Lorkowski S., Goel A., Ongen H., Green F., Clarke R., Collins R., Franzosi M.G., Tognoni G., et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17:806–814. doi:10.1093/hmg/ddm352. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

