Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity
- PMID: 22708079
- PMCID: PMC3375920
- DOI: 10.1016/j.celrep.2012.04.003
Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity
Abstract
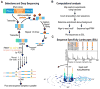
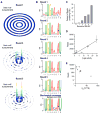
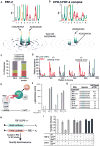
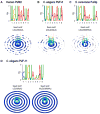
The control and function of RNA are governed by the specificity of RNA binding proteins. Here, we describe a method for global unbiased analysis of RNA-protein interactions that uses in vitro selection, high-throughput sequencing, and sequence-specificity landscapes. The method yields affinities for a vast array of RNAs in a single experiment, including both low- and high-affinity sites. It is reproducible and accurate. Using this approach,we analyzed members of the PUF (Pumilio and FBF) family of eukaryotic mRNA regulators. Our data identify effects of a specific protein partner on PUF-RNA interactions, reveal subsets of target sites not previously detected, and demonstrate that designer PUF proteins can precisely alter specificity. The approach described here is, in principle, broadly applicable for analysis of any molecule that binds RNA, including proteins, nucleic acids, and small molecules.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- 5T32HG002760/HG/NHGRI NIH HHS/United States
- R01 CA133508/CA/NCI NIH HHS/United States
- CA133508/CA/NCI NIH HHS/United States
- R01 GM050942/GM/NIGMS NIH HHS/United States
- GM050942/GM/NIGMS NIH HHS/United States
- F32 GM095169/GM/NIGMS NIH HHS/United States
- GM069420/GM/NIGMS NIH HHS/United States
- GM031892/GM/NIGMS NIH HHS/United States
- R37 GM031892/GM/NIGMS NIH HHS/United States
- T32 HG002760/HG/NHGRI NIH HHS/United States
- GM53320/GM/NIGMS NIH HHS/United States
- R01 GM069420/GM/NIGMS NIH HHS/United States
- R01 GM031892/GM/NIGMS NIH HHS/United States
- R01 GM053320/GM/NIGMS NIH HHS/United States
- GM095169/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

