Mitochondrial protein acetylation regulates metabolism
- PMID: 22708561
- PMCID: PMC3872051
- DOI: 10.1042/bse0520023
Mitochondrial protein acetylation regulates metabolism
Abstract
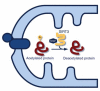
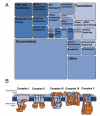
Changes in cellular nutrient availability or energy status induce global changes in mitochondrial protein acetylation. Over one-third of all proteins in the mitochondria are acetylated, of which the majority are involved in some aspect of energy metabolism. Mitochondrial protein acetylation is regulated by SIRT3 (sirtuin 3), a member of the sirtuin family of NAD+-dependent protein deacetylases that has recently been identified as a key modulator of energy homoeostasis. In the absence of SIRT3, mitochondrial proteins become hyperacetylated, have altered function, and contribute to mitochondrial dysfunction. This chapter presents a review of the functional impact of mitochondrial protein acetylation, and its regulation by SIRT3.
Figures
References
-
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. - PubMed
-
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. - PubMed
-
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

