Evolution of coding and non-coding genes in HOX clusters of a marsupial
- PMID: 22708672
- PMCID: PMC3541083
- DOI: 10.1186/1471-2164-13-251
Evolution of coding and non-coding genes in HOX clusters of a marsupial
Abstract
Background: The HOX gene clusters are thought to be highly conserved amongst mammals and other vertebrates, but the long non-coding RNAs have only been studied in detail in human and mouse. The sequencing of the kangaroo genome provides an opportunity to use comparative analyses to compare the HOX clusters of a mammal with a distinct body plan to those of other mammals.
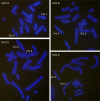
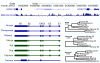
Results: Here we report a comparative analysis of HOX gene clusters between an Australian marsupial of the kangaroo family and the eutherians. There was a strikingly high level of conservation of HOX gene sequence and structure and non-protein coding genes including the microRNAs miR-196a, miR-196b, miR-10a and miR-10b and the long non-coding RNAs HOTAIR, HOTAIRM1 and HOXA11AS that play critical roles in regulating gene expression and controlling development. By microRNA deep sequencing and comparative genomic analyses, two conserved microRNAs (miR-10a and miR-10b) were identified and one new candidate microRNA with typical hairpin precursor structure that is expressed in both fibroblasts and testes was found. The prediction of microRNA target analysis showed that several known microRNA targets, such as miR-10, miR-414 and miR-464, were found in the tammar HOX clusters. In addition, several novel and putative miRNAs were identified that originated from elsewhere in the tammar genome and that target the tammar HOXB and HOXD clusters.
Conclusions: This study confirms that the emergence of known long non-coding RNAs in the HOX clusters clearly predate the marsupial-eutherian divergence 160 Ma ago. It also identified a new potentially functional microRNA as well as conserved miRNAs. These non-coding RNAs may participate in the regulation of HOX genes to influence the body plan of this marsupial.
Figures



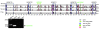



Similar articles
-
Molecular leveraging of HOX-embedded non-coding RNAs in the progression of acute myeloid leukemia.Hum Cell. 2024 Nov 30;38(1):24. doi: 10.1007/s13577-024-01149-9. Hum Cell. 2024. PMID: 39614990 Review.
-
Evolution of conserved non-coding sequences within the vertebrate Hox clusters through the two-round whole genome duplications revealed by phylogenetic footprinting analysis.J Mol Evol. 2010 Dec;71(5-6):427-36. doi: 10.1007/s00239-010-9396-1. Epub 2010 Oct 28. J Mol Evol. 2010. PMID: 20981416
-
Unique small RNA signatures uncovered in the tammar wallaby genome.BMC Genomics. 2012 Oct 17;13:559. doi: 10.1186/1471-2164-13-559. BMC Genomics. 2012. PMID: 23075437 Free PMC article.
-
Comparative analyses of vertebrate posterior HoxD clusters reveal atypical cluster architecture in the caecilian Typhlonectes natans.BMC Genomics. 2010 Nov 24;11:658. doi: 10.1186/1471-2164-11-658. BMC Genomics. 2010. PMID: 21106068 Free PMC article.
-
In silico interaction of HOX cluster-embedded microRNAs and long non-coding RNAs in oral cancer.J Oral Pathol Med. 2022 Jan;51(1):18-29. doi: 10.1111/jop.13225. Epub 2021 Aug 16. J Oral Pathol Med. 2022. PMID: 34358375 Review.
Cited by
-
The function of homeobox genes and lncRNAs in cancer.Oncol Lett. 2016 Sep;12(3):1635-1641. doi: 10.3892/ol.2016.4901. Epub 2016 Jul 21. Oncol Lett. 2016. PMID: 27588114 Free PMC article.
-
Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia.Tumour Biol. 2016 Aug;37(8):10571-6. doi: 10.1007/s13277-016-4948-7. Epub 2016 Feb 9. Tumour Biol. 2016. PMID: 26857279
-
HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy.Mol Cell Biochem. 2017 Aug;432(1-2):179-187. doi: 10.1007/s11010-017-3008-y. Epub 2017 Mar 18. Mol Cell Biochem. 2017. PMID: 28316060
-
Expression and clinicopathological significance of the lncRNA HOXA11-AS in colorectal cancer.Oncol Lett. 2016 Nov;12(5):4155-4160. doi: 10.3892/ol.2016.5129. Epub 2016 Sep 14. Oncol Lett. 2016. PMID: 27895785 Free PMC article.
-
MiR-196b Promotes the Invasion and Migration of Lung Adenocarcinoma Cells by Targeting AQP4.Technol Cancer Res Treat. 2021 Jan-Dec;20:1533033820985868. doi: 10.1177/1533033820985868. Technol Cancer Res Treat. 2021. PMID: 33455522 Free PMC article.
References
-
- Papageorgiou S. HOX Gene Expression. Landes Bioscience, Texas; 2007.
-
- Pourquié O. HOX Genes. Elsevier, San Diego; 2009.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

