Genomic characterization of explant tumorgraft models derived from fresh patient tumor tissue
- PMID: 22709571
- PMCID: PMC3439334
- DOI: 10.1186/1479-5876-10-125
Genomic characterization of explant tumorgraft models derived from fresh patient tumor tissue
Abstract
Background: There is resurgence within drug and biomarker development communities for the use of primary tumorgraft models as improved predictors of patient tumor response to novel therapeutic strategies. Despite perceived advantages over cell line derived xenograft models, there is limited data comparing the genotype and phenotype of tumorgrafts to the donor patient tumor, limiting the determination of molecular relevance of the tumorgraft model. This report directly compares the genomic characteristics of patient tumors and the derived tumorgraft models, including gene expression, and oncogenic mutation status.
Methods: Fresh tumor tissues from 182 cancer patients were implanted subcutaneously into immune-compromised mice for the development of primary patient tumorgraft models. Histological assessment was performed on both patient tumors and the resulting tumorgraft models. Somatic mutations in key oncogenes and gene expression levels of resulting tumorgrafts were compared to the matched patient tumors using the OncoCarta (Sequenom, San Diego, CA) and human gene microarray (Affymetrix, Santa Clara, CA) platforms respectively. The genomic stability of the established tumorgrafts was assessed across serial in vivo generations in a representative subset of models. The genomes of patient tumors that formed tumorgrafts were compared to those that did not to identify the possible molecular basis to successful engraftment or rejection.
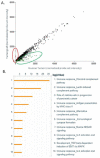
Results: Fresh tumor tissues from 182 cancer patients were implanted into immune-compromised mice with forty-nine tumorgraft models that have been successfully established, exhibiting strong histological and genomic fidelity to the originating patient tumors. Comparison of the transcriptomes and oncogenic mutations between the tumorgrafts and the matched patient tumors were found to be stable across four tumorgraft generations. Not only did the various tumors retain the differentiation pattern, but supporting stromal elements were preserved. Those genes down-regulated specifically in tumorgrafts were enriched in biological pathways involved in host immune response, consistent with the immune deficiency status of the host. Patient tumors that successfully formed tumorgrafts were enriched for cell signaling, cell cycle, and cytoskeleton pathways and exhibited evidence of reduced immunogenicity.
Conclusions: The preservation of the patient's tumor genomic profile and tumor microenvironment supports the view that primary patient tumorgrafts provide a relevant model to support the translation of new therapeutic strategies and personalized medicine approaches in oncology.
Figures




References
-
- Fiebig HH, Schuchhardt C, Henss H, Fiedler L, Lohr GW. Comparison of tumor response in nude mice and in the patients. Behring Inst Mitt. 1984;74:343–352. - PubMed
-
- Garber K. From human to mouse and back: 'tumorgraft' models surge in popularity. J Natl Cancer Inst. 2009;101:6–8. - PubMed
-
- Fiebig HH, Widmer K-H, Fiedler L, WIttekind C, Lohr GW. Development and Characterization of 51 Human Tumor Models for Large Bowel, Stomach and Esophageal Cancers. Dig Surg. 1984;1:225–235. doi: 10.1159/000171659. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

