Broadening the spectrum of β-lactam antibiotics through inhibition of signal peptidase type I
- PMID: 22710113
- PMCID: PMC3421906
- DOI: 10.1128/AAC.00726-12
Broadening the spectrum of β-lactam antibiotics through inhibition of signal peptidase type I
Abstract
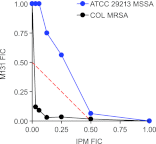
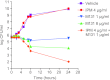
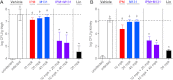
The resistance of methicillin-resistant Staphylococcus aureus (MRSA) to all β-lactam classes limits treatment options for serious infections involving this organism. Our goal is to discover new agents that restore the activity of β-lactams against MRSA, an approach that has led to the discovery of two classes of natural product antibiotics, a cyclic depsipeptide (krisynomycin) and a lipoglycopeptide (actinocarbasin), which potentiate the activity of imipenem against MRSA strain COL. We report here that these imipenem synergists are inhibitors of the bacterial type I signal peptidase SpsB, a serine protease that is required for the secretion of proteins that are exported through the Sec and Tat systems. A synthetic derivative of actinocarbasin, M131, synergized with imipenem both in vitro and in vivo with potent efficacy. The in vitro activity of M131 extends to clinical isolates of MRSA but not to a methicillin-sensitive strain. Synergy is restricted to β-lactam antibiotics and is not observed with other antibiotic classes. We propose that the SpsB inhibitors synergize with β-lactams by preventing the signal peptidase-mediated secretion of proteins required for β-lactam resistance. Combinations of SpsB inhibitors and β-lactams may expand the utility of these widely prescribed antibiotics to treat MRSA infections, analogous to β-lactamase inhibitors which restored the utility of this antibiotic class for the treatment of resistant Gram-negative infections.
Figures



Similar articles
-
Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics.Sci Transl Med. 2012 Mar 21;4(126):126ra35. doi: 10.1126/scitranslmed.3003592. Sci Transl Med. 2012. PMID: 22440737
-
Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA.Nat Chem Biol. 2015 Nov;11(11):855-61. doi: 10.1038/nchembio.1911. Epub 2015 Sep 14. Nat Chem Biol. 2015. PMID: 26368589 Free PMC article.
-
Sensitizing of β-lactam resistance by tannic acid in methicillin-resistant S. aureus.World J Microbiol Biotechnol. 2019 Mar 21;35(4):57. doi: 10.1007/s11274-019-2637-6. World J Microbiol Biotechnol. 2019. PMID: 30900046
-
Auxiliary factors: a chink in the armor of MRSA resistance to β-lactam antibiotics.Curr Opin Microbiol. 2013 Oct;16(5):538-48. doi: 10.1016/j.mib.2013.06.012. Epub 2013 Jul 26. Curr Opin Microbiol. 2013. PMID: 23895826 Review.
-
Can β-Lactam Antibiotics Be Resurrected to Combat MRSA?Trends Microbiol. 2019 Jan;27(1):26-38. doi: 10.1016/j.tim.2018.06.005. Epub 2018 Jul 18. Trends Microbiol. 2019. PMID: 30031590 Review.
Cited by
-
Systems-level antimicrobial drug and drug synergy discovery.Nat Chem Biol. 2013 Apr;9(4):222-31. doi: 10.1038/nchembio.1205. Nat Chem Biol. 2013. PMID: 23508188
-
Prospects for Antibacterial Discovery and Development.J Am Chem Soc. 2021 Dec 22;143(50):21127-21142. doi: 10.1021/jacs.1c10200. Epub 2021 Dec 3. J Am Chem Soc. 2021. PMID: 34860516 Free PMC article.
-
Stable Signal Peptides and the Response to Secretion Stress in Staphylococcus aureus.mBio. 2017 Dec 12;8(6):e01507-17. doi: 10.1128/mBio.01507-17. mBio. 2017. PMID: 29233892 Free PMC article.
-
Unrealized targets in the discovery of antibiotics for Gram-negative bacterial infections.Nat Rev Drug Discov. 2023 Dec;22(12):957-975. doi: 10.1038/s41573-023-00791-6. Epub 2023 Oct 13. Nat Rev Drug Discov. 2023. PMID: 37833553 Review.
-
Emerging peptide antibiotics with therapeutic potential.Med Drug Discov. 2021 Mar;9:100078. doi: 10.1016/j.medidd.2020.100078. Epub 2020 Dec 30. Med Drug Discov. 2021. PMID: 33398258 Free PMC article. Review.
References
-
- Becher D, et al. 2009. A proteomic view of an important human pathogen—towards the quantification of the entire Staphylococcus aureus proteome. PLoS One 4:e8176 doi:10.1371/journal.pone.0008176 - DOI - PMC - PubMed
-
- Berger-Bächi B, Rohrer S. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165–171 - PubMed
-
- Bonapace CR, White RL, Friedrich LV, Bosso JA. 2000. Evaluation of antibiotic synergy against Acinetobacter baumannii: a comparison with Etest, time-kill, and checkerboard methods. Diagn. Microbiol. Infect. Dis. 38:43–50 - PubMed
-
- Bruton G, et al. 2003. Lipopeptide substrates for SpsB, the Staphylococcus aureus type I signal peptidase: design, conformation and conversion to α-ketoamide inhibitors. Eur. J. Med. Chem. 38:351–356 - PubMed
-
- Buzder-Lantos P, Bockstael K, Anné J, Herdewijn P. 2009. Substrate based peptide aldehyde inhibits bacterial type I signal peptidase. Bioorg. Med. Chem. Lett. 19:2880–2883 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

