TGF-β control of stem cell differentiation genes
- PMID: 22710171
- PMCID: PMC3466472
- DOI: 10.1016/j.febslet.2012.03.023
TGF-β control of stem cell differentiation genes
Abstract
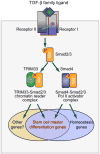
The canonical TGF-β/Smad signaling pathway was delineated in the mid 90s and enriched over the past decade with many findings about its specificity, regulation, networking, and malfunctions in disease. However, a growing understanding of the chromatin status of a critical class of TGF-β target genes - the genes controlling differentiation of embryonic stem cells - recently prompted a reexamination of this pathway and its critical role in the regulation of stem cell differentiation. The new findings reveal master regulators of the pluripotent state set the stage for Smad-mediated activation of master regulators of the next differentiation stage. Furthermore, a novel branch of the TGF-β/Smad pathway has been identified in which a chromatin-reading Smad complex makes the master differentiation genes accessible to canonical Smad complexes for transcriptional activation. These findings provide exciting new insights into the global role of TGF-β signaling in the regulators of stem cell fate.
Copyright © 2012 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1gamma to Chromatin via Its PHD Finger-Bromodomain Activates Its Ubiquitin Ligase and Transcriptional Repressor Activities. Mol Cell. 2011;43:85–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

