N-acetylgalactosamine utilization pathway and regulon in proteobacteria: genomic reconstruction and experimental characterization in Shewanella
- PMID: 22711537
- PMCID: PMC3431674
- DOI: 10.1074/jbc.M112.382333
N-acetylgalactosamine utilization pathway and regulon in proteobacteria: genomic reconstruction and experimental characterization in Shewanella
Abstract
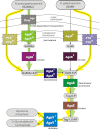
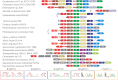
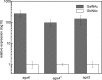
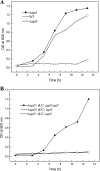
We used a comparative genomics approach to reconstruct the N-acetyl-d-galactosamine (GalNAc) and galactosamine (GalN) utilization pathways and transcriptional regulons in Proteobacteria. The reconstructed GalNAc/GalN utilization pathways include multiple novel genes with specific functional roles. Most of the pathway variations were attributed to the amino sugar transport, phosphorylation, and deacetylation steps, whereas the downstream catabolic enzymes in the pathway were largely conserved. The predicted GalNAc kinase AgaK, the novel variant of GalNAc-6-phosphate deacetylase AgaA(II) and the GalN-6-phosphate deaminase AgaS from Shewanella sp. ANA-3 were validated in vitro using individual enzymatic assays and reconstitution of the three-step pathway. By using genetic techniques, we confirmed that AgaS but not AgaI functions as the main GalN-6-P deaminase in the GalNAc/GalN utilization pathway in Escherichia coli. Regulons controlled by AgaR repressors were reconstructed by bioinformatics in most proteobacterial genomes encoding GalNAc pathways. Candidate AgaR-binding motifs share a common sequence with consensus CTTTC that was found in multiple copies and arrangements in regulatory regions of aga genes. This study provides comprehensive insights into the common and distinctive features of the GalNAc/GalN catabolism and its regulation in diverse Proteobacteria.
Figures

References
-
- Freymond P. P., Lazarevic V., Soldo B., Karamata D. (2006) Poly(glucosyl-N-acetylgalactosamine 1-phosphate), a wall teichoic acid of Bacillus subtilis 168: its biosynthetic pathway and mode of attachment to peptidoglycan. Microbiology 152, 1709–1718 - PubMed
-
- Bernatchez S., Szymanski C. M., Ishiyama N., Li J., Jarrell H. C., Lau P. C., Berghuis A. M., Young N. M., Wakarchuk W. W. (2005) A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J. Biol. Chem. 280, 4792–4802 - PubMed
-
- Carraway K. L., Hull S. R. (1991) Cell surface mucin-type glycoproteins and mucin-like domains. Glycobiology 1, 131–138 - PubMed
-
- Abu-Qarn M., Eichler J., Sharon N. (2008) Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18, 544–550 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

