Associating microbiome composition with environmental covariates using generalized UniFrac distances
- PMID: 22711789
- PMCID: PMC3413390
- DOI: 10.1093/bioinformatics/bts342
Associating microbiome composition with environmental covariates using generalized UniFrac distances
Abstract
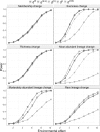
Motivation: The human microbiome plays an important role in human disease and health. Identification of factors that affect the microbiome composition can provide insights into disease mechanism as well as suggest ways to modulate the microbiome composition for therapeutical purposes. Distance-based statistical tests have been applied to test the association of microbiome composition with environmental or biological covariates. The unweighted and weighted UniFrac distances are the most widely used distance measures. However, these two measures assign too much weight either to rare lineages or to most abundant lineages, which can lead to loss of power when the important composition change occurs in moderately abundant lineages.
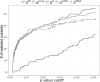
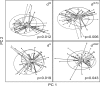
Results: We develop generalized UniFrac distances that extend the weighted and unweighted UniFrac distances for detecting a much wider range of biologically relevant changes. We evaluate the use of generalized UniFrac distances in associating microbiome composition with environmental covariates using extensive Monte Carlo simulations. Our results show that tests using the unweighted and weighted UniFrac distances are less powerful in detecting abundance change in moderately abundant lineages. In contrast, the generalized UniFrac distance is most powerful in detecting such changes, yet it retains nearly all its power for detecting rare and highly abundant lineages. The generalized UniFrac distance also has an overall better power than the joint use of unweighted/weighted UniFrac distances. Application to two real microbiome datasets has demonstrated gains in power in testing the associations between human microbiome and diet intakes and habitual smoking.
Availability: http://cran.r-project.org/web/packages/GUniFrac
Figures


References
-
- Carr D., et al. hexbin: Hexagonal Binning Routines. 2011
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

