Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species
- PMID: 22711811
- PMCID: PMC3390827
- DOI: 10.1073/pnas.1201403109
Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species
Abstract
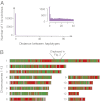
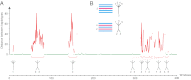
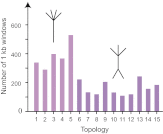
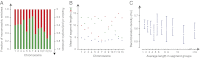
In a genome alignment of five individuals of the ascomycete fungus Zymoseptoria pseudotritici, a close relative of the wheat pathogen Z. tritici (synonym Mycosphaerella graminicola), we observed peculiar diversity patterns. Long regions up to 100 kb without variation alternate with similarly long regions of high variability. The variable segments in the genome alignment are organized into two main haplotype groups that have diverged ∼3% from each other. The genome patterns in Z. pseudotritici are consistent with a hybrid speciation event resulting from a cross between two divergent haploid individuals. The resulting hybrids formed the new species without backcrossing to the parents. We observe no variation in 54% of the genome in the five individuals and estimate a complete loss of variation for at least 30% of the genome in the entire species. A strong population bottleneck following the hybridization event caused this loss of variation. Variable segments in the Z. pseudotritici genome exhibit the two haplotypes contributed by the parental individuals. From our previously estimated recombination map of Z. tritici and the size distribution of variable chromosome blocks untouched by recombination we estimate that the hybridization occurred ∼380 sexual generations ago. We show that the amount of lost variation is explained by genetic drift during the bottleneck and by natural selection, as evidenced by the correlation of presence/absence of variation with gene density and recombination rate. The successful spread of this unique reproductively isolated pathogen highlights the strong potential of hybridization in the emergence of pathogen species with sexual reproduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. - PubMed
-
- Greig D, Louis EJ, Borts RH, Travisano M. Hybrid speciation in experimental populations of yeast. Science. 2002;298:1773–1775. - PubMed
-
- Lexer C, Welch ME, Durphy JL, Rieseberg LH. Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: Implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol Ecol. 2003;12:1225–1235. - PubMed
-
- Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

